Figure 4.
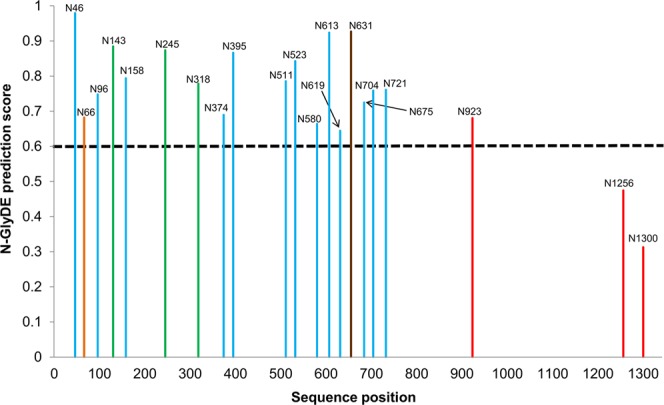
N-GlyDE prediction results of human VEGFR2, where sites with a prediction score above 0.6 (shown by the dotted line) are predicted as glycosites. Green bars represent the three glycosites with experimental evidence in UniProt. Glycosites annotated by sequence analysis in UniProt were recently validated by Chandler et al.’s mass spectrometry (MS) experiment on extracellular domain of murine VEGFR2, which shares 86% sequence similarity with human VEGFR2. Blue bars represent the MS-validated glycosites; the brown bar (N631) represents sites undetected in the MS experiment; and the orange bar (N66) represents the sequon only in human, not in murine. Red bars represent the non-glycosites, which was not studied by Chandler et al. The number following ‘N’ represents the asparagine position of the sequon in the sequence.
