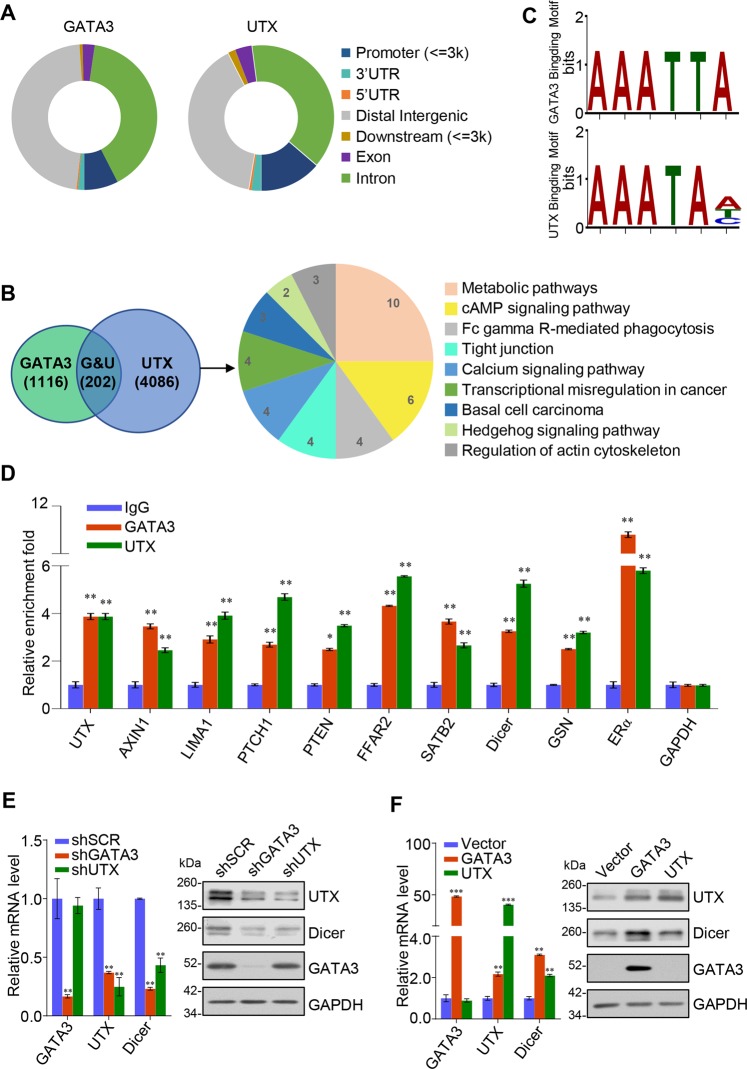
Fig. 5. Genome-wide transcriptional-target analysis for GATA3/UTX/MLL4 complex.
a Genomic distribution of GATA3 and UTX targets, based on ChIP-seq analysis. b Left: Venn diagram showing overlapping promoters bound by GATA3 and UTX in MCF-7 cells. Right: pathway analysis of 202 overlapping target genes of GATA3 and UTX arranged into functional groups. c MEME-ChIP analysis of GATA3- and UTX-bound motifs. d Verification of ChIP-seq results through qChIP analysis of indicated genes in MCF-7 cells. Results are expressed as fold-change relative to control, and GAPDH was used as a negative control. Data are shown as means ± SD from three independent experiments. e qPCR and western blotting analysis of the expression of UTX and Dicer in GATA3- or UTX-depleted MCF-7 cells. f qPCR and western blotting analysis of the expression of UTX and Dicer in GATA3- or UTX-overexpressing MDA-MB-231 cells. d–f Two-tailed unpaired t test (*p < 0.05, **p < 0.01, ***p < 0.001)