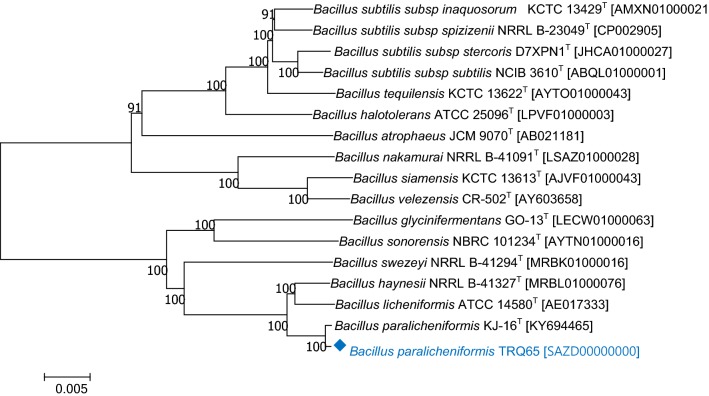
Fig. 1.
Phylogenetic relation between TRQ65 and closely related species: Bacillus paralicheniformis KJ-16T [KY694465]; Bacillus haynesii NRRL B-41327T [MRBL01000076]; Bacillus licheniformis ATCC 14580T [AE017333]; Bacillus glycinifermentans GO-13T [LECW01000063]; Bacillus sonorensis NBRC 101234T [AYTN01000016]; Bacillus swezeyi NRRL B-41294T [MRBK01000096]; Bacillus subtilis subsp. spizizenii NRRL B-23049T [CP002905]; Bacillus subtilis subsp. inaquosorum KCTC 13429T [AMXN01000021]; Bacillus tequilensis KCTC 13622T [AYTO01000043]; Bacillus subtilis subsp. subtilis NCIB 3610T [ABQL01000001]; Bacillus halotolerans ATCC 25096T [LPVF01000003]; Bacillus subtilis subsp. stercoris D7XPN1T [JHCA01000027]; Bacillus atrophaeus JCM 9070T [AB021181]; Bacillus nakamurai NRRL B-41091T [LSAZ01000028]; Bacillus siamensis KCTC 13613T [AJVF01000043]; Bacillus velezensis CR-502T [AY603658], constructed by the builder method in REALPHY 1.12 (Bertels et al. 2014) and MEGA 7 (Kumar et al. 2016) using the neighbor-joining algorithm (based on 1000 bootstrap replications). Scale bar (0.005) represents the number of nucleotide substitutions per site