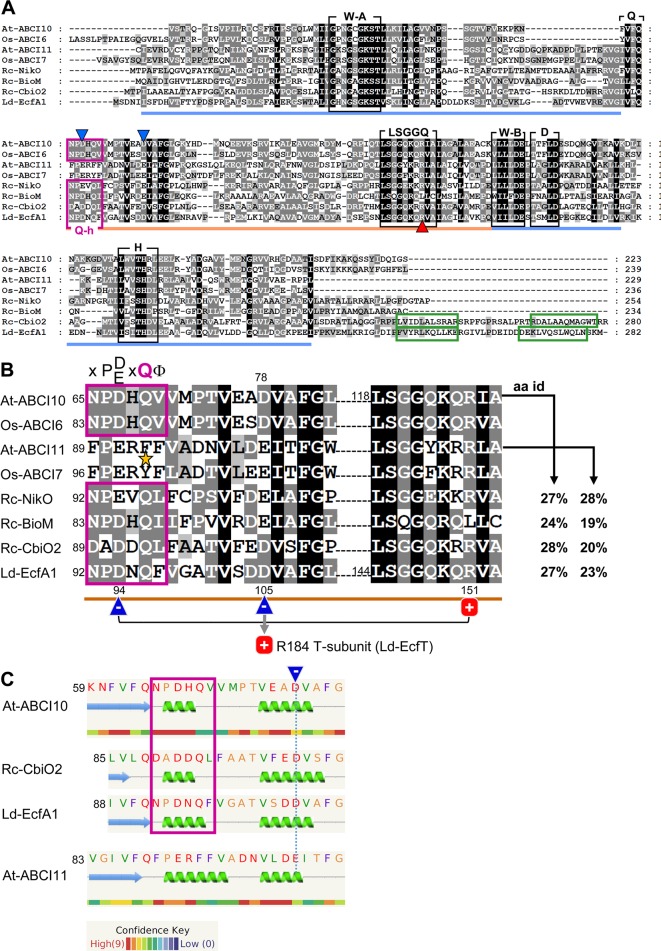
Figure 1.
ABCI10 and ABCI11 proteins. (A) Amino acid sequences of mature Arabidopsis At-ABCI10 (Q8H1R4), At-ABCI11 (Q8LEF6), rice Os-ABCI6 (Q5ZD09), Os-ABCI7 (Q2R434), and the bacterial ECF transporter A subunits NikO (D5AQY6), BioM (D5ARH0), CbiO2 (O68106) from Rhodobacter capsulatus and EcfA1 (Q1GBJ0) from Lactobacillus delbrueckii. UniProtKB accession numbers are given in brackets (UniProt Consortium, 2018), and chloroplast targeting peptides were predicted by ChloroP (Emanuelsson et al., 1999). Please note that in rice, the protein Os-ABCI7 (Q2R434) was named Os-ABCI8 by Zeng et al. (2017). The conserved motifs Walker A, Walker B, Q-loop, D-loop, and H-loop (black boxes) form the nucleotide binding sites in the RecA domain (blue line). The ABC-transporter ATPase specific helical subdomain (orange line) contains the signature motif LSGGQ. The Q-helix (purple box) and the two α-helices in the C-terminal domain of CbiO2 and EcfA1 (green boxes) are indicated. Conserved negative acidic residues are highlighted by blue, and the conserved positively charged arginines are highlighted by red triangles. All motifs and domains are depicted according to the crystal structures of Rc-CbiO2 and Ld-EcfA1 in the CbiMQO and FolT2 complexes (pdb entries 5X3X and 5D7T). (B) Highlighted Q-helix (purple box) and helical subdomain region from alignment in (A). In ECF transporter A subunits, the Q-helix with the specific motif X-P-D/E-X-Q-Φ (X is any, Φ a hydrophobic amino acid) directly follows the Q-loop that connects the RecA and helical subdomain [see (A)]. The invariant glutamine (Q) residue of the Q-helix is absent in ABCI11 proteins (asterisk). According to the crystal structure of FolT2 (Swier et al., 2016; Rempel et al., 2019), the conserved acidic and negatively charged residue (blue triangle) in the Q-helix of EcfA1 (D94) forms an intramolecular interaction with the conserved positive arginine (R151) in the LSGGQ motif (red square). Further, the invariant acidic residue in the first helix of the helical subdomain (D105 of LcfA1) is positioned at the surface of the negatively charged groove of the EcfA1A2 dimer and binds to the conserved arginine (R184) in the coupling helix 2 of the membrane-intrinsic EcfT subunit (compare Supplementary Figure 10 ). The amino acid identities (aa id) of the full mature At-ABCI10 and At-ABCI11 proteins to Rc-NikO, Rc-BioM, Rc-CbiO2, and Ld-EcfA1 are indicated. (C) Secondary structure and disorder prediction by Phyre2 (Kelley et al., 2015) reveals that the Q-helix predicted for At-ABCI10 most likely forms a short helical turn specific for ECF transporter ATPase subunits like CbiO2 and EcfA1. In contrast, substitution of the conserved polar glutamine (Q) by a hydrophobic phenylalanine (F) in At-ABCI11 leads to a less confident prediction of a longer helix. In consequence, the acidic glutamate in the first α-helix of the helical subdomain of At-ABCI11 (dotted blue line) might not be positioned for proper interaction with residues of a membrane-intrinsic T-subunit.