Figure 1.
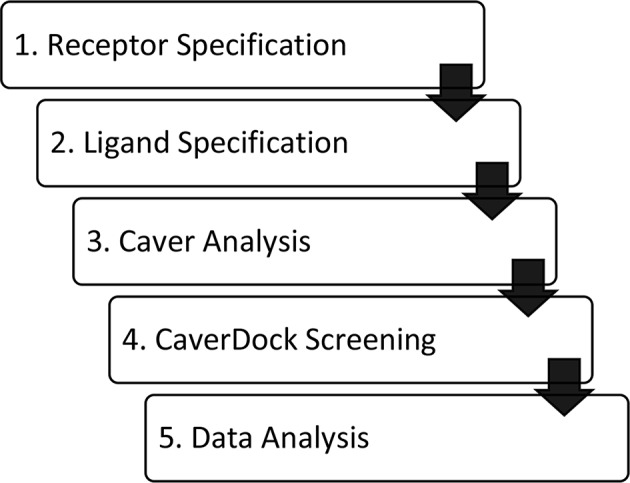
Workflow of virtual screening using CaverDock. (1) Receptor and (2) ligand specification follow established protocols of the software tool AutoDock (Morris et al., 2009). (3) Identification of protein tunnels using Caver (Chovancova et al., 2012). (4) Tunnel discretization and sequential ligand binding study using CaverDock (Filipovic et al., 2019; Vavra et al., 2019). (5) Analysis of docking trajectories and energy profiles, extraction of energy barriers and protein-ligand complexes possessing the lowest energies.
