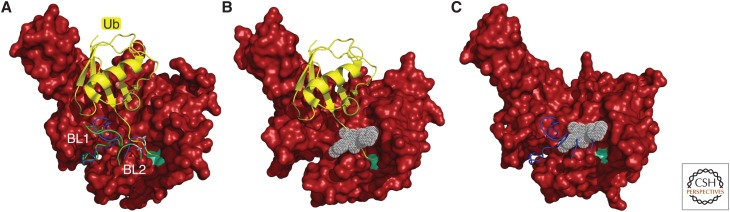
Figure 6.
Activation and inhibition of USP14. (A) Structure of the substrate-engaged USP14 (red) bound to ubiquitin aldehyde (Ub-Al, in yellow), a model of the enzyme's activated state. The principal difference from the ubiquitin-free (inactive) state of USP14 is in the positioning of the two blocking (BL) loops (Hu et al. 2005). The BL loops of USP14-Ub-Al are in green, those of the free USP14 in blue. To the right of these loops, the active-site Cys is represented as a light green patch. In the free form of USP14, the BL loops block the access of the carboxyl terminus of ubiquitin to the active site. This occlusion is relieved in the substrate-engaged form. Ser432 (gray), located within the BL2 loop, is subject to phosphorylation by AKT, providing partial activation (Xu et al. 2015). (B) Structure of USP14 bound to IU1-47 (white) shows that this inhibitor occludes access of the carboxyl terminus of ubiquitin to the catalytic site (Wang et al. 2018). To better visualize the IU1-47-binding pocket, both BL loops were removed from the density maps. (C) Structure of free USP14 with IU1-47 superimposed. This model shows how the BL2 loop (blue) occludes access of IU1-47, consistent with previous evidence that IU1 series compounds do not inhibit the free form of USP14 (Lee et al. 2016). Density maps of the USP14 catalytic domain in its free form (Protein Data Bank [PDB]: 2AYN), USP14 bound to Ub-Al (PDB: 2AYO), and the USP14-IU1-47 complex (PDB: 6IIL) were created from data in Hu et al. (2005) and Wang et al. (2018). To better visualize the catalytic site of USP14, Gln197 was removed from all USP14 structures in this figure.