Abstract
RNA silencing pathways play critical roles in maintaining quiescence of transposons in germ cells to promote genome integrity. However the precise mechanism by which different types of transposons are recognized by these pathways is not fully understood. Furthermore, the location in the germline where this transposition occurs after disruption of transposon silencing was previously unknown. Here we utilize the spatial and temporal organization of the Caenorhabditis elegans germline to demonstrate that transposition of DNA transposons in RNA silencing pathway mutants occur in all stages of adult germ cells. We further demonstrate that the double-strand breaks generated by transposons can restore homologous recombination in a mutant defective for the generation of meiosis-specific double-strand breaks. Finally, we detected clear differences in transposase expression and transposon excision between distinct branches of the RNA silencing pathway, emphasizing that there are multiple mechanisms by which transposons can be recognized and routed for small-RNA-mediated silencing.
Keywords: transposons, double-strand breaks, RNAi, germline, C . elegans
Transposons are discrete segments of DNA that are capable of excising themselves from one locus and reintegrating themselves at another genomic location. Movement of transposons in and out of genes can alter their expression and function, making transposons a major source of deleterious mutations as well as a driving force of evolution. In many organisms, transposons have also been co-opted by researchers for mapping, random and site-directed mutagenesis, and gene tagging (Williams et al. 1992; Barrett et al. 2004; Williams et al. 2005; Robert and Bessereau 2007; Frøkjaer-Jensen et al. 2008; Frøkjær-Jensen et al. 2010). Because transposons utilize their host’s cellular machinery for their mobilization, they are considered to be selfish DNA parasites, similar to viruses.
There are two major classes of transposable elements – retrotransposons (Class I), which contain an open reading frame coding for a retroviral-like reverse transcriptase and transpose through an RNA intermediate, and DNA transposons (Class II), which move via a DNA-based “cut-and-paste” mechanism. DNA transposons usually contain a transposase sequence flanked by Terminal Inverted Repeats (TIRs). The transposase recognizes the specific sequence of its TIRs and catalyzes a cleavage reaction that releases the transposon ends. The transposase also recognizes a preferred target site, and inserts the transposon at the chosen location (Bessereau 2006). At the site of excision, a DNA transposon leaves behind a double-strand break (DSB), which must be repaired by the host’s cellular machinery, either through homologous recombination or non-homologous end joining. The mechanism of repair is determined primarily based on cell type – somatic cells favor end joining pathways whereas germ cells often repair breaks via homologous recombination, and a subset of these events are resolved as interhomolog crossovers (Plasterk 1991; Robert et al. 2008).
There are numerous retrotransposons in the genome, which, until recently, were thought to be inactive. However, a study published in 2012 demonstrated that CER1, Gypsy-like retrotransposon, is transcriptionally active and produces viral-like particles in wild-type C. elegans germlines (Dennis et al. 2012). More recently, it has been demonstrated that several other retrotransposons, including CER3, are targets of the nuclear RNA interference (RNAi) pathway and H3K9 methylation (Ni et al. 2014; 2016; Zeller et al. 2016; Ni et al. 2018). It is not yet known whether any of these retrotransposons are capable of transposition in C. elegans. In contrast, transposition has been detected for at least seven distinct families of DNA transposons (Tc1-Tc5, Tc7, CemaT1), though there are many more transposons present that have not been well studied (Eide and Anderson 1985; Collins et al. 1989; Levitt and Emmons 1989; Yuan et al. 1991; Collins and Anderson 1994; Rezsohazy et al. 1997; Brownlie and Whyard 2004; Bessereau 2006). The most well characterized DNA transposon family in C. elegans is Tc1, of which there are 31 intact copies present in the genome (Fischer et al. 2003). Tc1 is not normally active in germ cells, however, gene mutations that result in activation of Tc1 were identified from a forward genetic screen and are referred to as mutator (mut) class genes (Ketting et al. 1999). Around the same time, a screen for mutations that result in defects in RNAi identified a largely overlapping panel of genes, suggesting that the silencing of transposons is an endogenous function of the RNAi pathway (Tabara et al. 1999).
Many of the mutator pathway genes have been identified as components of the small RNA-mediated silencing pathways, including the nucleotidyl transferase (mut-2/rde-3), the 3′-5′ exonuclease (mut-7), the DEAD box RNA helicase (mut-14), the glutamine/asparagine (Q/N)-rich protein (mut-16/rde-6), two proteins of unknown function (mut-8/rde-2 and mut-15), (Ketting et al. 1999; Tijsterman et al. 2002; Vastenhouw et al. 2003; Tops et al. 2005; Chen et al. 2005; Gu et al. 2009). C. elegans with mutations in these genes not only have active transposons and defects in response to exogenous RNAi, but also have temperature-sensitive sterility and defects in endogenous siRNA production (Gu et al. 2009; Zhang et al. 2011; Phillips et al. 2014). All of proteins encoded by these mutator pathway genes, along with the RNA-dependent RNA polymerase RRF-1, associate to form a protein complex that synthesizes highly abundant secondary 22G-siRNAs (22 nucleotides starting with a 5′ guanosine) that function downstream of primary Argonaute proteins (Pak and Fire 2007; Sijen et al. 2007; Gu et al. 2009; Gent et al. 2010; Phillips et al. 2012). This complex forms nuclear pore-associated perinuclear condensates in germ cells, referred to as Mutator foci, where it is thought to play a key role in surveillance and silencing of deleterious transcripts, including transposon-derived RNAs, as they exit the nucleus (Phillips et al. 2012; Uebel et al. 2018).
In addition to endogenous siRNAs, PIWI-associated small RNAs (piRNAs) also have roles in silencing transposons (Batista et al. 2008; Das et al. 2008). In C. elegans, piRNAs (also referred to as 21U-RNAs) are bound and stabilized by the PIWI protein PRG-1 and trigger production of secondary 22G-siRNAs dependent on the mutator pathway (Ruby et al. 2006; Wang and Reinke 2008; Batista et al. 2008; Das et al. 2008; Lee et al. 2012; Bagijn et al. 2012). Only a single transposon family, Tc3, has been demonstrated to transpose upon loss of the piRNA machinery (Das et al. 2008), however, multiple other DNA transposons are up-regulated transcriptionally or lose mutator pathway-dependent 22G-siRNAs (Bagijn et al. 2012; McMurchy et al. 2017).
Here we demonstrate that DSBs generated by transposition of DNA transposons in mutator pathway or piRNA pathway mutants can be visualized throughout the germline of adult C. elegans, allowing us to determine both temporally and spatially where these transposons are active. Furthermore, in mutator pathway mutants these transposon-mediated DSBs can, at some frequency, be repaired by homologous recombination. Thus mutator pathway mutants can partially rescue the meiotic defects of spo-11 mutants, which fail to initiate meiotic recombination through the generation of DSBs. Finally, we observe distinct differences in transposon mRNA expression and frequency of DSBs generated by transposition between mutator pathway and piRNA pathway mutants, highlighting the distinct roles these two pathways play in transposon silencing.
Materials and Methods
C. elegans strains
Unless otherwise stated, worms were grown at 20° according to standard conditions (Brenner 1974). Strains used in this study include:
RNA isolation and quantitative RT-PCR
RNA was isolated from synchronized adult C. elegans (66-68 h after L1 arrest) using Trizol, followed by chloroform extraction and isopropanol precipitation. qRT-PCR was performed using transposon-specific primer pairs and rpl-32 for normalization (Table 1). Data were analyzed using the 2-ΔΔCt method and P-values were calculated in R using the t-test function in the package ‘pcr’ (Ahmed and Kim 2018).
Table 1. Primers used in this study.
| Name | Sequence |
|---|---|
| Tc1 - F | TGGGCTAAACACATCTGGTC |
| Tc1 - R | CGGTTGGGCATTGATACTTTG |
| Tc2 - F | AGTTATGAGGATTGGATGGTGC |
| Tc2 - R | AGTATTGGAGCATTGACGGC |
| Tc3 - F | GTCCGTATCGTGTATGCTCAG |
| Tc3 - R | AATAGACTTCCAAGCGTCGAG |
| Tc4v - F | GTAATCGCTGAACCAAAAGGC |
| Tc4v - R | GTGTCTTGTATCCAGCCCG |
| Tc5 - F | AGTGTACCGTGTCTTTCGTG |
| Tc5 - R | GGAGTTTCCACTTTGACATGTTG |
| RTE1 - F | CCCTGGAATGAGAGTGAATGG |
| RTE1 - R | GTACGAGTTCTTGGAGCATTTTG |
| CER1/Gypsy - F | CCCGGAACTATGCTCATTCTAG |
| CER1/Gypsy - R | TCAGTACAGACGAAGCAGTTC |
| Mirage - F | AGAAGCTGAAACCGATGAGTC |
| Mirage - R | TCAGAGAACGACACAGTTGAC |
| rpl-32 - F | CAAGGTCGTCAAGAAGAAGC |
| rpl-32 - R | GGCTACACGACGGTATCTGT |
Fluorescent microscopy
C. elegans were picked as L4s and dissected the following day for immunofluorescence for most experiments. For diakinesis imaging and scoring, animals were picked as L4 and kept for three days at 15° prior to dissection. All strains carrying the spo-11 mutation were selected as L4s from the progeny of balanced spo-11/nT1 animals. Gonads were immunostained according to previously described protocol with rabbit anti-RAD-51 (SDIX, 2948.00.02), guinea pig anti-HTP-3 (MacQueen et al. 2005), and Alexa Fluor secondary antibodies purchased from ThermoFisher (Phillips et al. 2009). Imaging was performed on an Axio Imager Z1 microscope with ApoTome running Axiovision software (Zeiss) or a DeltaVision Elite microscope running SoftWoRx (GE Healthcare). Images were collected as three-dimensional data stacks, displayed as maximum intensity projections, and pseudocolored using Adobe Photoshop.
RAD-51 quantification
Age-matched (one day post-L4) hermaphrodite gonads were immunostained for RAD-51 and imaged. Gonads were divided into six zones of equal length, starting at the distal tip through the end of pachytene and the number of foci per nucleus were scored for each zone. Three gonads were scored for each genotype.
Brood size analysis
Hermaphrodites of the indicated genotypes were placed on individual plates as L4-stage larvae. They were moved to fresh plates approximately every 24 hr until egg laying was complete. At the time the animal was removed from the plate, the total number of embryos and hatched larvae was counted. Approximately three days later, the total number of hermaphrodite and male progeny on the plate was scored. Total number of broods scored was 11 broods for wild-type, 29 broods for spo-11, 13 broods for mut-7; spo-11, 15 broods for mut-16; spo-11, and 21 broods for prg-1; spo-11.
Recombination analysis
Hermaphrodites heterozygous for dpy-3unc-3 and homozygous for spo-11 were generated by mating balanced spo-11/mIs11 males to mIs11; dpy-3unc-3 hermaphrodites. The transgene mIs11 is located on chromosome IV near the spo-11 locus; it can be identified by a pharyngeal GFP signal and was used to balance the spo-11 mutant. The spo-11/mIs11; dpy-3unc-3 F2 hermaphrodites were then mated to spo-11/mIs11 males to generate the spo-11; dpy-3unc-3/++ strain used for recombination analysis. The cross was performed similarly using mut-16/+; spo-11/mIs11 males and mut-16; mIs11; dpy-3unc-3 hermaphrodites to generate the mut-16; spo-11; dpy-3unc-3/++ and the mut-16/+; spo-11; dpy-3unc-3/++ strains, which were identified as homozygous or heterozygous for mut-16 by genotyping after egg-laying was completed. Progeny from each cross were scored as wild-type, Unc Dpy, Unc non-Dpy, or Dpy non-Unc, and as hermaphrodite or male. Recombination frequency (p) for hermaphrodites was calculated as P = 1 – , where R is the fraction of recombinant progeny, scored as two times the number of Unc non-Dpy hermaphrodites to account for the possibility that Dpy non-Unc animals are non-recombinant triplo-X animals (Brenner 1974). For males, recombination frequency is calculated as P = R. The total recombination frequency is calculated ((2 * phermaphrodite * [# of hermaphrodites]) + (pmale * [# of males]))/(2 * [# of hermaphrodites] + [# of males]), which accounts for hermaphrodites having two X chromosomes and males only one (Kelly et al. 2000). Map distances in cM = 100 × p. Total number of broods scored was 28 broods for spo-11; dpy-3 unc-3/++, 7 broods for mut-16/+; spo-11; dpy-3 unc-3/++, and 9 broods for mut-16; spo-11; dpy-3 unc-3/++.
Data Availability
All strains are available either at the Caenorhabditis Genetics Center (CGC) or upon request from the Phillips lab. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Results
Transposon mRNA expression profiles of RNAi pathway mutants
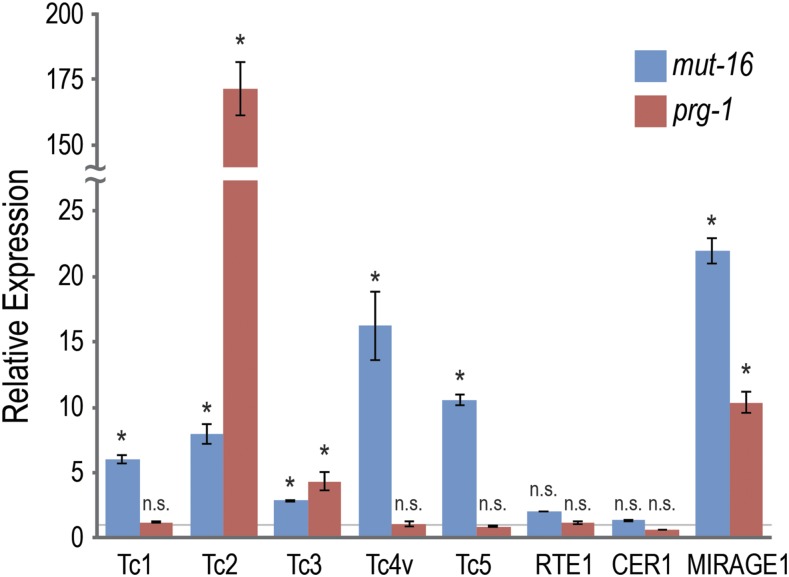
It is well known that RNAi pathways regulate DNA transposon activity in the C. elegans germline (Billi et al. 2014). However, the specific temporal and spatial region of the germline where this transposition occurs has not previously been studied. To address this question, we sought to visualize transposon activity by utilizing the DSBs left behind by the DNA transposons when they transpose, which are then repaired by cellular DNA repair machinery. First, however, we sought to determine how transposon activity differs between distinct branches of the RNAi pathway. It was previously reported that mutants in the mutator pathway and the piRNA pathway have different rates of transposon mobilization depending on the transposon being examined. For example, Tc1, Tc3, and Tc4 transposons are active in a mut-7 mutant, whereas only the Tc3 transposon is active in a prg-1 mutant (Das et al. 2008). As a preliminary analysis of which transposons may mobilize in mutants from either the mutator pathway or the piRNA pathway, we performed qRT-PCR analysis of transposon mRNA expression in mut-16 and prg-1 mutants (Figure 1). Specifically, we examined the mRNA expression from several known DNA transposons (Tc1, Tc2, Tc3, Tc4v, Tc5, and MIRAGE1) and two retrotransposons (RTE1 and CER1/Gypsy) and found that, of the DNA transposons, Tc1, Tc4v and Tc5 had significantly increased mRNA expression in mut-16 but not prg-1 mutants, whereas Tc2, Tc3, and MIRAGE1 had increased expression in both mutants. Interestingly, Tc2 was significantly higher in prg-1 (170-fold) compared to mut-16 (eightfold), which is surprising because piRNA-mediated silencing is generally thought to be upstream of and to require the mutator pathway (Das et al. 2008; Lee et al. 2012; Bagijn et al. 2012). It is important to note that this analysis is only indicative of transposon mRNA expression in the RNAi pathway mutants relative to wild-type animals, and is not direct evidence of transposon mobilization rates. Furthermore, while this analysis does not distinguish between somatic and germline transposon activity, it does suggest that Tc2 transposon silencing may be mediated, at least in part, by a piRNA pathway that is independent of the mutator pathway and WAGO 22G-siRNAs.
Figure 1.
Mutator pathway mutants and piRNA pathway mutants have distinct profiles of transposon mRNA expression. Primers recognizing the transposon mRNAs were used for quantitative RT-PCR with rpl-32 as a normalization standard. Expression levels shown are relative to wild-type animals (gray horizontal line) and error bars represent the standard deviation of two technical replicates. Two primer sets were used for each transposon mRNA with similar results, however only one representative set is shown. n.s denotes not significant and indicates p-value > 0.05 and * indicates p-value < 0.05.
Visualization and quantification of transposon mobilization
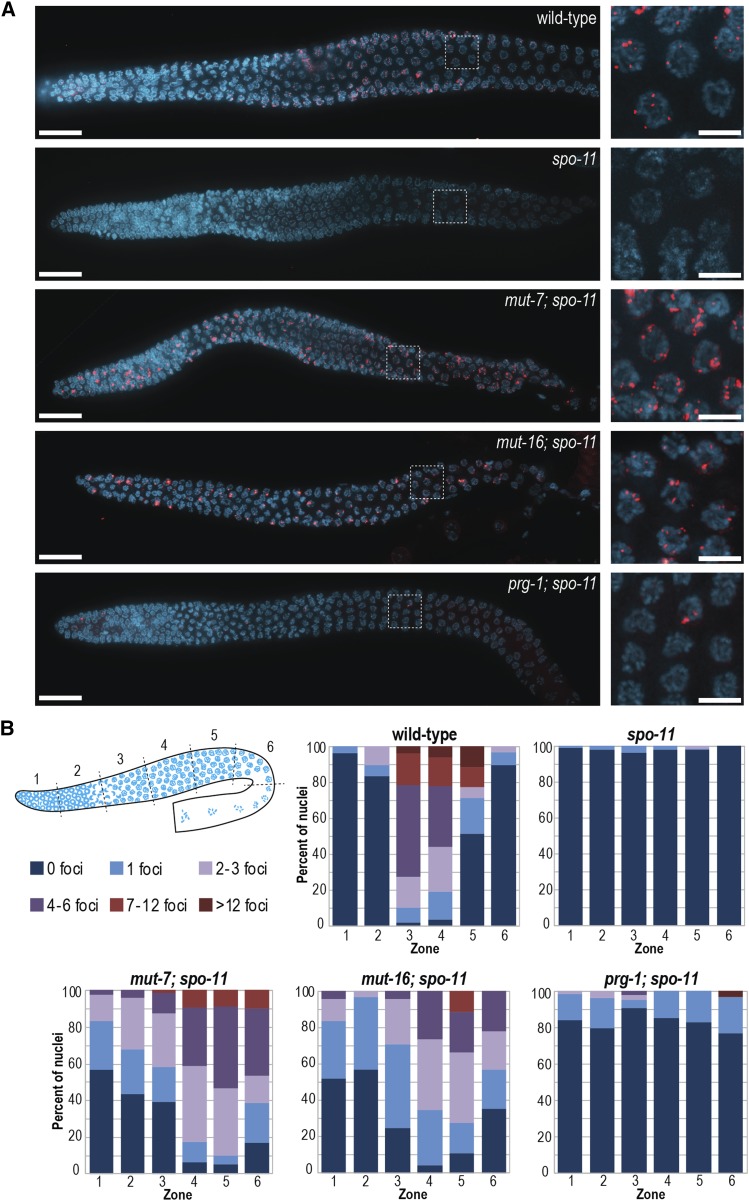
To visualize transposon activity specifically in the C. elegans germline, we chose to examine the expression of RAD-51, a homolog of the bacterial RecA protein and a key protein in DSB repair pathways (Ogawa et al. 1993). In wild-type C. elegans, RAD-51 can be visualized as distinct, punctate foci in the zygotene/pachytene stages of meiosis (Figure 2A) (Alpi et al. 2003). We initially examined multiple mutants in the RNAi pathway for increased RAD-51 foci in germ cells, however, the presence of programmed DSBs generated for meiotic recombination, complicated the analysis. To alleviate this problem, we crossed the RNAi pathway mutants into a spo-11 mutant. SPO-11 is the type-II topoisomerase that is required to initiate meiotic recombination through the generation of DSBs (Keeney et al. 1997; Dernburg et al. 1998). In the spo-11 mutant, RAD-51 foci are virtually eliminated (Figure 2A-B) (Alpi et al. 2003), providing us a background where we can examine spo-11-independent DSBs generated due to transposon mobilization. We first examined the germline of mutator mutants (mut-7 or mut-16) in the spo-11 background. In these strains we could visualize numerous DSBs throughout the germline, starting in the mitotic proliferation zone, and extending through the meiotic stages of leptotene, zygotene, pachytene, and diplotene (Figure 2A-B). Because the number of foci increases as the nuclei progress through the meiotic program in the mutator pathway mutants, we can infer that transposons are generating new DSBs throughout these stages. Additionally, we examined the germline of C. elegans with a mutation in the piRNA pathway (prg-1). In prg-1 mutants we observed significantly fewer DSBs compared to the mutator mutants, but higher levels than in the spo-11 mutant alone (Figure 2A-B). These results are consistent with a role for the piRNA pathway in silencing only a subset of transposons, in contrast to the mutator pathway, which is more broadly required for transposon silencing.
Figure 2.
SPO-11-independent RAD-51 foci are present throughout the germlines of mutator pathway mutants. (A) Whole gonads (left) stained with RAD-51 (red) and DAPI (blue). RAD-51 foci can be seen throughout the germlines of mut-7; spo-11, mut-16; spo-11, and to a lesser extent, prg-1; spo-11. This is in contrast to wild-type, where the majority of RAD-51 foci are found in the zygotene to mid-pachytene region. Scale bars, 20μm. Magnification of pachytene stage nuclei (right) stained with RAD-51 (red) and DAPI (blue). Scale bars, 5μm. (B) Diagram (top left) depicting the six zones in which RAD-51 foci were quantified. Stacked bar charts show percent of nuclei in each zone with the specified number of RAD-51 foci for the indicated genotypes. X axes indicate the position in the germline (zone).
Transposon-induced DSBs can partially rescue meiotic defects of spo-11 mutants
We next sought to examine whether transposon-induced DSBs are competent to rescue the meiotic phenotypes of the spo-11 mutant. In the absence of functional SPO-11 protein, chromosomes fail to undergo meiotic recombination (Keeney et al. 1997; Dernburg et al. 1998). Failure to undergo meiotic recombination causes errors in segregation of chromosomes at the meiosis I division, ultimately resulting in aneuploidy and embryonic lethality. The few progeny surviving to adulthood from spo-11 mutants are frequently males (Dernburg et al. 1998). This “High incidence of males” or Him phenotype is also indicative of a chromosome segregation defect; male C. elegans have a single sex chromosome (XO) and thus mis-segregation of the X chromosomes in an XX hermaphrodite results in an increased production of males (Hodgkin et al. 1979). In contrast to the spo-11 null mutants, which largely produce inviable embryos that fail to survive to adulthood (2.8% viable), mut-7; spo-11 and mut-16; spo-11 produce 12.1% and 10.7% viable embryos that survive to adulthood, respectively (Table 2). Similarly, mut-7; spo-11 and mut-16; spo-11 produce fewer male progeny (14.9% males for mut-7; spo-11 and 21.4% males for mut-16; spo-11) than the spo-11 mutant alone (40.9% males) (Table 2). Unlike the mutator pathway mutants, the piRNA pathway mutant prg-1 failed to rescue embryonic viability or the production of male progeny. These data indicate that the DSBs generated by transposon mobilization in the mutator pathway mutants, but not piRNA pathway mutants, can compensate for the lack of SPO-11 protein and increase the frequency of proper chromosome segregation, presumably by promoting the formation of crossovers.
Table 2. Mutator pathway mutations increase progeny viability and reduce the number of self-progeny males in a spo-11 mutant.
| Genotype | % Viable Embryosa | % Male Progenyb |
|---|---|---|
| wild-type | 100.00 (n = 3035) | 0.07 (n = 3035) |
| spo-11 | 2.80 (n = 5328) | 40.94 (n = 149) |
| mut-7; spo-11 | 12.11 (n = 1882) | 14.91 (n = 228) |
| mut-16; spo-11 | 10.68 (n = 2144) | 21.40 (n = 229) |
| prg-1; spo-11 | 2.12 (n = 4105) | 51.72 (n = 87) |
Total number of embryos scored to calculate % viable embryos indicated in parentheses.
Total number of adults scored to calculate % male self-progeny indicated in parentheses.
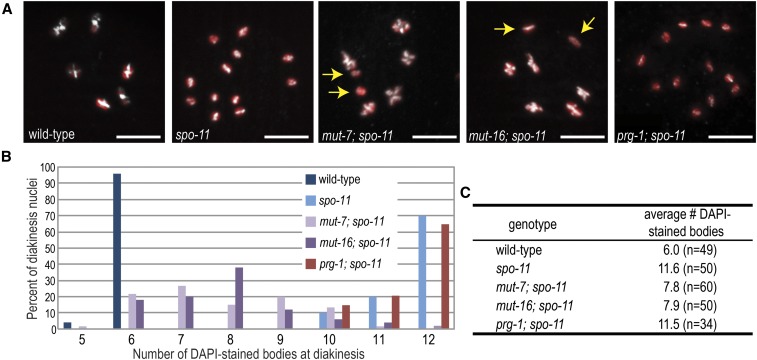
To test directly whether DSBs generated by transposon mobilization can promote the formation of crossovers, we examined diakinesis stage of meiosis for the presence of recombinant chromosomes. In wild-type C. elegans, six bivalents (pairs of recombinant chromosomes) are present, whereas, in spo-11, 12 non-recombinant univalents can be observed (Villeneuve 1994; Dernburg et al. 1998). The mutator pathway mutants, mut-7 and mut-16, were able to partially rescue the spo-11 diakinesis phenotype, averaging approximately eight DAPI-staining bodies, but with a range of six to 11 DAPI-staining bodies (Figure 3A-C). In contrast, the piRNA pathway mutant, prg-1, was indistinguishable from the spo-11 mutant alone with 12 univalents (Figure 3A-C). We also analyzed the frequency of recombination between two genetic markers, dpy-3 and unc-3, which lie on opposite ends of the X chromosome, a distance of ∼38 cM in wild-type animals (Villeneuve 1994; Dernburg et al. 1998; Kelly et al. 2000). In spo-11 mutants, recombination is undetectable in this region (Dernburg et al. 1998) whereas mut-16; spo-11 mutants we calculated the map distance between dpy-3 and unc-3 as 26.6 cM (∼70% of wild-type) (Table 3). These data indicate that, in each germline nucleus of a mutator pathway mutant, most chromosomes have at least one mobilized transposon generating a DSB that is subsequently repaired by homologous recombination. Because some DSBs are occurring well before or after the stage at which nuclei are competent for homologous recombination and because many DSBs may be repaired by other mechanisms, these figures significantly underestimate the total number of mobilized transposons per nucleus.
Figure 3.
Mutations in the mutator pathway can restore crossover formation in the spo-11 mutant. (A) Representative wild-type and mutant diakinesis oocytes stained with HTP-3 (white) and DAPI (red) to allow for counting of the number of the number of bivalents (homologous chromosomes connected by chiasmata) or univalents in each strain. Yellow arrows in mut-7; spo-11 and mut-16; spo-11 point to a single pair of non-recombinant chromosomes. Scale bars, 5μm. (B) Graph indicating the number of DAPI-stained bodies in diakinesis oocytes for each genotype. Wild-type oocytes display six DAPI-stained bivalents, representing the six pairs of chromosomes held together by chiasmata, while spo-11, which fails to make double-strand breaks for recombination, displays 12 DAPI-stained univalents. Mutations in the mutator pathway but not the piRNA pathway can partially rescue the spo-11 phenotype. Occasionally, two bivalents lie too close together to be visually resolved, resulting in a modest underestimation of the number of DAPI-stained bodies. (C) Mean number of DAPI-stained bodies scored for each of the genotypes in (B). Total number of oocytes scored is indicated in parentheses.
Table 3. A mutation in the mutator pathway restores recombination in the spo-11 mutant.
| Recombinant hermaphrodites | Recombinant males | ||||||
|---|---|---|---|---|---|---|---|
| Genotype | Unc non-Dpy hermaphrodites | Dpy non-Unc hermaphrodites | Total hermaphrodites | Unc non-Dpy males | Dpy non-Unc males | Total males | Map distance (cM)a |
| +/+ or mut-16/+; spo-11; dpy-3 unc-3/++ | 0 | 0 | 134 | 0 | 0 | 131 | 0.0 |
| mut-16; spo-11; dpy-3 unc-3/++ | 27 | 29 | 239 | 9 | 10 | 60 | 26.6 |
Map distance was calculated as described in Materials and Methods.
Discussion
By visualizing transposon-derived DSBs as RAD-51 foci present in a spo-11 mutant background, we can provide quantification of transposon-hopping levels in the mutator pathway and piRNA pathway mutant backgrounds. Furthermore, in mutator mutants, transposon-derived DSBs can rescue spo-11 mutant phenotypes, including recombination frequency, chiasma formation, viability, and male production. The assays described could be extended to examine new mutants in the transposon-silencing pathway, to screen for new mutants by taking advantage of the increased fertility of spo-11 mutants when combined with mutations in the transposon silencing pathway, or to probe more deeply into which classes of transposons are mobilized and the frequency using ChIP-seq of RAD-51.
Interestingly, we observe clear differences in the expression of transposon mRNAs by qRT-PCR and in the rates of transposition assayed by frequency of DSBs, demonstrating that these two pathways do not have fully overlapping roles in transposon silencing. This result, along with previously reported differences in Tc1 and Tc4 mobilization between the two pathways (Das et al. 2008), is somewhat surprising because piRNA pathways are thought to be the primary mediator of transposon silencing in many organisms (Czech and Hannon 2016). That leads to the question of how transposons silenced independently of piRNAs are recognized. piRNA-targeting can trigger multigenerational silencing that can be maintained in the absence of the initial piRNA trigger (Ashe et al. 2012; Shirayama et al. 2012; Luteijn et al. 2012). Thus one possibility is that silencing of these transposons was initiated by piRNAs, but when those piRNAs were lost, silencing was maintained by mutator-dependent heritable siRNAs. In fact, a mutation in hrde-1, the Argonaute protein required to inherit siRNAs from one generation to the next, in combination with a mutation in prg-1, desilences the Tc1 transposon to a level similar to that of a mutation in the mutator pathway (de Albuquerque et al. 2015). Alternatively, some features of the transposon mRNA structure could be recognized by the cell as aberrant and routed for silencing completely independently of piRNAs. For example, multiple reports have implicated splicing factors in the RNA silencing pathway, suggesting that irregular introns or mis-spliced mRNAs can be a signal for siRNA-mediated silencing (Kim et al. 2005; Dumesic et al. 2013; Akay et al. 2017; Tyc et al. 2017; Newman et al. 2018). Many C. elegans transposons contain introns, including Tc1 and Tc3 (van Luenen et al. 1993; Vos et al. 1993). Interestingly, the Tc1 intron is inefficiently spliced, yet 22G-siRNAs are exclusively made from spliced transcripts which accumulate in spliceosomes, suggesting that siRNA biogenesis is downstream of splicing-mediated surveillance (Sijen and Plasterk 2003; Newman et al. 2018). How these surveillance mechanisms are interwoven to mediate efficient recognition of transposon and other foreign mRNAs remains to be determined.
Acknowledgments
We thank the members of the Phillips lab for helpful discussions and feedback on the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and Shohei Mitani of the Japanese National BioResource Project. This work was supported in part by a Basil O’Connor Starter Scholar Research Award from the March of Dimes Foundation Grant No. 5-FY17-38 (to C.M.P.), the National Science Foundation Graduate Research Fellowship Program Grant No. DGE 1418060 (to C.J.U.), and the National Institute of Heath Grants T32 GM118289 (to D.H.N.), K22 CA177897 (to C.M.P.) and R35 GM119656 (to C.M.P.). C.M.P is a Pew Scholar in the Biomedical Sciences supported by the Pew Charitable Trusts and C.J.U. is a USC Dornsife-funded Chemistry-Biology Interface trainee. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Communicating editor: J. Kim
Literature Cited
- Ahmed M., and Kim D. R., 2018. pcr: an R package for quality assessment, analysis and testing of qPCR data. PeerJ 6: e4473 10.7717/peerj.4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akay A., Di Domenico T., Suen K. M., Nabih A., Parada G. E. et al. , 2017. The Helicase Aquarius/EMB-4 Is Required to Overcome Intronic Barriers to Allow Nuclear RNAi Pathways to Heritably Silence Transcription. Dev. Cell 42: 241–255.e6. 10.1016/j.devcel.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi A., Pasierbek P., Gartner A., and Loidl J., 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16. 10.1007/s00412-003-0237-5 [DOI] [PubMed] [Google Scholar]
- Ashe A., Sapetschnig A., Weick E.-M., Mitchell J., Bagijn M. P. et al. , 2012. piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell 150: 88–99. 10.1016/j.cell.2012.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn M. P., Goldstein L. D., Sapetschnig A., Weick E.-M., Bouasker S. et al. , 2012. Function, Targets, and Evolution of Caenorhabditis elegans piRNAs. Science 337: 574–578. 10.1126/science.1220952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett P. L., Fleming J. T., and Göbel V., 2004. Targeted gene alteration in Caenorhabditis elegans by gene conversion. Nat. Genet. 36: 1231–1237. 10.1038/ng1459 [DOI] [PubMed] [Google Scholar]
- Batista P. J., Ruby J. G., Claycomb J. M., Chiang R., Fahlgren N. et al. , 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31: 67–78. 10.1016/j.molcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessereau J.-L., 2006. Transposons in C. elegans. WormBook, ed. The C. elegans Research Community WormBook, 10.1895/wormbook.1.70.1, http://www.wormbook.org. [DOI] [Google Scholar]
- Billi A. C., Fischer S. E. J., and Kim J. K., 2014. Endogenous RNAi pathways in C. elegans. WormBook, ed. The C. elegans Research Community WormBook, http://www.wormbook.org. 10.1895/wormbook.1.170.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie J. C., and Whyard S., 2004. CemaT1 is an active transposon within the Caenorhabditis elegans genome. Gene 338: 55–64. 10.1016/j.gene.2004.05.011 [DOI] [PubMed] [Google Scholar]
- Chen C.-C. G., Simard M. J., Tabara H., Brownell D. R., McCollough J. A. et al. , 2005. A member of the polymerase beta nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr. Biol. 15: 378–383. 10.1016/j.cub.2005.01.009 [DOI] [PubMed] [Google Scholar]
- Collins J., Forbes E., and Anderson P., 1989. The Tc3 family of transposable genetic elements in Caenorhabditis elegans. Genetics 121: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. J., and Anderson P., 1994. The Tc5 family of transposable elements in Caenorhabditis elegans. Genetics 137: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B., and Hannon G. J., 2016. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem. Sci. 41: 324–337. 10.1016/j.tibs.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P. P., Bagijn M. P., Goldstein L. D., Woolford J. R., Lehrbach N. J. et al. , 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31: 79–90. 10.1016/j.molcel.2008.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque B. F. M., Placentino M., and Ketting R. F., 2015. Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans. Dev. Cell 34: 448–456. 10.1016/j.devcel.2015.07.010 [DOI] [PubMed] [Google Scholar]
- Dennis S., Sheth U., Feldman J. L., English K. A., and Priess J. R., 2012. C. elegans Germ Cells Show Temperature and Age-Dependent Expression of Cer1, a Gypsy/Ty3-Related Retrotransposon. PLoS Pathog. 8: e1002591 10.1371/journal.ppat.1002591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M. et al. , 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. 10.1016/S0092-8674(00)81481-6 [DOI] [PubMed] [Google Scholar]
- Dumesic P. A., Natarajan P., Chen C., Drinnenberg I. A., Schiller B. J. et al. , 2013. Stalled Spliceosomes Are a Signal for RNAi-Mediated Genome Defense. Cell 152: 957–968. 10.1016/j.cell.2013.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D., and Anderson P., 1985. Transposition of Tc1 in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 82: 1756–1760. 10.1073/pnas.82.6.1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S. E. J., Wienholds E., and Plasterk R. H. A., 2003. Continuous exchange of sequence information between dispersed Tc1 transposons in the Caenorhabditis elegans genome. Genetics 164: 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Hollopeter G., Taylor J., Harris T. W. et al. , 2010. Targeted gene deletions in C. elegans using transposon excision. Nat. Methods 7: 451–453. 10.1038/nmeth.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M. et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J. I., Lamm A. T., Pavelec D. M., Maniar J. M., Parameswaran P. et al. , 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell 37: 679–689. 10.1016/j.molcel.2010.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D., Vasale J., Batista P. J. et al. , 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36: 231–244. 10.1016/j.molcel.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., and Brenner S., 1979. Nondisjunction Mutants of the Nematode CAENORHABDITIS ELEGANS. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., and Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- Kelly K. O., Dernburg A. F., Stanfield G. M., and Villeneuve A. M., 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R. F., Haverkamp T. H., van Luenen H. G., and Plasterk R. H. A., 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99: 133–141. 10.1016/S0092-8674(00)81645-1 [DOI] [PubMed] [Google Scholar]
- Kim J. K., Gabel H. W., Kamath R. S., Tewari M., Pasquinelli A. E. et al. , 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167. 10.1126/science.1109267 [DOI] [PubMed] [Google Scholar]
- Lee H.-C., Gu W., Shirayama M., Youngman E., Conte D. et al. , 2012. C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. Cell 150: 78–87. 10.1016/j.cell.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt A., and Emmons S. W., 1989. The Tc2 transposon in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 86: 3232–3236. 10.1073/pnas.86.9.3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn M. J., van Bergeijk P., Kaaij L. J. T., Almeida M. V., Roovers E. F. et al. , 2012. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31: 3422–3430. 10.1038/emboj.2012.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Phillips C. M., Bhalla N., Weiser P., Villeneuve A. M. et al. , 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123: 1037–1050. 10.1016/j.cell.2005.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurchy A. N., Stempor P., Gaarenstroom T., Wysolmerski B., Dong Y. et al. , 2017. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. eLife 6: e21666 (erratum: eLife 6: 32516). 10.7554/eLife.21666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. A., Ji F., Fischer S. E. J., Anselmo A., Sadreyev R. I. et al. , 2018. The surveillance of pre-mRNA splicing is an early step in C. elegans RNAi of endogenous genes. Genes Dev. 32: 670–681. 10.1101/gad.311514.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Z., Chen E., and Gu S. G., 2014. Complex coding of endogenous siRNA, transcriptional silencing and H3K9 methylation on native targets of germline nuclear RNAi in C. elegans. BMC Genomics 15: 1157 10.1186/1471-2164-15-1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Z., Kalinava N., Chen E., Huang A., Trinh T. et al. , 2016. A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics Chromatin 9: 3 10.1186/s13072-016-0052-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Z., Kalinava N., Mendoza S. G., and Gu S. G., 2018. The spatial and temporal dynamics of nuclear RNAi-targeted retrotransposon transcripts in Caenorhabditis elegans. Development 145: dev167346. 10.1242/dev.167346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Yu X., Shinohara A., and Egelman E. H., 1993. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259: 1896–1899. 10.1126/science.8456314 [DOI] [PubMed] [Google Scholar]
- Pak J., and Fire A., 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244. 10.1126/science.1132839 [DOI] [PubMed] [Google Scholar]
- Phillips C. M., McDonald K. L., and Dernburg A. F., 2009. Cytological analysis of meiosis in Caenorhabditis elegans. Methods Mol. Biol. 558: 171–195. 10.1007/978-1-60761-103-5_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Montgomery B. E., Breen P. C., Roovers E. F., Rim Y.-S. et al. , 2014. MUT-14 and SMUT-1 DEAD Box RNA Helicases Have Overlapping Roles in Germline RNAi and Endogenous siRNA Formation. Curr. Biol. 24: 839–844. 10.1016/j.cub.2014.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Montgomery T. A., Breen P. C., and Ruvkun G., 2012. MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26: 1433–1444. 10.1101/gad.193904.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H. A., 1991. The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J. 10: 1919–1925. 10.1002/j.1460-2075.1991.tb07718.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezsohazy R., van Luenen H. G., Durbin R. M., and Plasterk R. H. A., 1997. Tc7, a Tc1-hitch hiking transposon in Caenorhabditis elegans. Nucleic Acids Res. 25: 4048–4054. 10.1093/nar/25.20.4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V. J., and Bessereau J.-L., 2007. Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 26: 170–183. 10.1038/sj.emboj.7601463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V. J., Davis M. W., Jorgensen E. M., and Bessereau J.-L., 2008. Gene conversion and end-joining-repair double-strand breaks in the Caenorhabditis elegans germline. Genetics 180: 673–679. 10.1534/genetics.108.089698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J. G., Jan C. H., Player C., Axtell M. J., Lee W. et al. , 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207. 10.1016/j.cell.2006.10.040 [DOI] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H.-C., Gu W., Ishidate T. et al. , 2012. piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell 150: 65–77. 10.1016/j.cell.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., and Plasterk R. H. A., 2003. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426: 310–314. 10.1038/nature02107 [DOI] [PubMed] [Google Scholar]
- Sijen T., Steiner F. A., Thijssen K. L., and Plasterk R. H. A., 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247. 10.1126/science.1136699 [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian M., Kelly W. G., Fleenor J., Grishok A. et al. , 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132. 10.1016/S0092-8674(00)81644-X [DOI] [PubMed] [Google Scholar]
- Tijsterman M., Ketting R. F., Okihara K. L., Sijen T., and Plasterk R. H. A., 2002. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science 295: 694–697. 10.1126/science.1067534 [DOI] [PubMed] [Google Scholar]
- Tops B. B. J., Tabara H., Sijen T., Simmer F., Mello C. C. et al. , 2005. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res. 33: 347–355. 10.1093/nar/gki183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K. M., Nabih A., Wu M. Z., Wedeles C. J., Sobotka J. A. et al. , 2017. The Conserved Intron Binding Protein EMB-4 Plays Differential Roles in Germline Small RNA Pathways of C. elegans. Dev. Cell 42: 256–270.e6. 10.1016/j.devcel.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Uebel C. J., Anderson D. C., Mandarino L. M., Manage K. I., Aynaszyan S. et al. , 2018. Distinct regions of the intrinsically disordered protein MUT-16 mediate assembly of a small RNA amplification complex and promote phase separation of Mutator foci. PLoS Genet. 14: e1007542 10.1371/journal.pgen.1007542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Luenen H. G., Colloms S. D., and Plasterk R. H. A., 1993. Mobilization of quiet, endogenous Tc3 transposons of Caenorhabditis elegans by forced expression of Tc3 transposase. EMBO J. 12: 2513–2520. 10.1002/j.1460-2075.1993.tb05906.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw N. L., Fischer S. E. J., Robert V. J., Thijssen K. L., Fraser A. G. et al. , 2003. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr. Biol. 13: 1311–1316. 10.1016/S0960-9822(03)00539-6 [DOI] [PubMed] [Google Scholar]
- Villeneuve A. M., 1994. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136: 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos J. C., van Luenen H. G., and Plasterk R. H. A., 1993. Characterization of the Caenorhabditis elegans Tc1 transposase in vivo and in vitro. Genes Dev. 7: 1244–1253. 10.1101/gad.7.7a.1244 [DOI] [PubMed] [Google Scholar]
- Wang G., and Reinke V., 2008. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 18: 861–867. 10.1016/j.cub.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. D., Schrank B., Huynh C., Shownkeen R., and Waterston R. H., 1992. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics 131: 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. C., Boulin T., Ruaud A.-F., Jorgensen E. M., and Bessereau J.-L., 2005. Characterization of Mos1-mediated mutagenesis in Caenorhabditis elegans: a method for the rapid identification of mutated genes. Genetics 169: 1779–1785. 10.1534/genetics.104.038265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. Y., Finney M., Tsung N., and Horvitz H. R., 1991. Tc4, a Caenorhabditis elegans transposable element with an unusual fold-back structure. Proc. Natl. Acad. Sci. USA 88: 3334–3338. 10.1073/pnas.88.8.3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P., Padeken J., van Schendel R., Kalck V., Tijsterman M. et al. , 2016. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48: 1385–1395. 10.1038/ng.3672 [DOI] [PubMed] [Google Scholar]
- Zhang C., Montgomery T. A., Gabel H. W., Fischer S. E. J., Phillips C. M. et al. , 2011. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108: 1201–1208. 10.1073/pnas.1018695108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains are available either at the Caenorhabditis Genetics Center (CGC) or upon request from the Phillips lab. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.