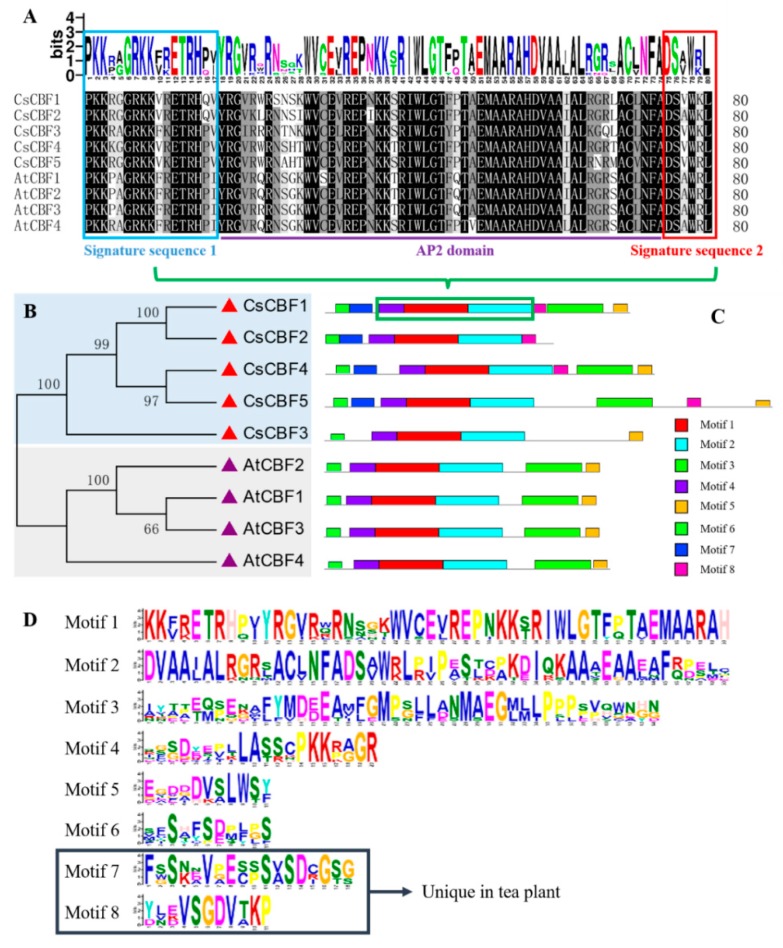
Figure 1.
Sequence alignment, phylogenetic and conserved motif analysis of CsCBFs and AtCBFs. (A) Multiple sequence alignment of the AP2 DNA-binding domain and two signature sequences and generating a logo. The Y axis (measured in bits) of logo depicts the overall height of the stack, indicating the sequence conservation at that position, while the height of symbols within the stack indicates the relative frequency of each amino at that position. The purple underline indicates the AP2 DNA-binding domain; the blue box represents the PKK/RPAGRxKFxETRHP signature sequence, and the red box represents the DSAWR signature sequence. (B) Phylogenetic analysis of the CBF protein sequences from tea plants and Arabidopsis. The red triangle represents the tea plants and the purple triangle represents the Arabidopsis. The phylogenetic tree was constructed using MEGA 5.0 software and the Neighbor-Joining method. The number above/below the branch represents the bootstrap values (from 1000 replicates). (C) Eight conserved motifs were predicted by MEME. (D) Detailed information of the eight conserved motifs.