Table 1.
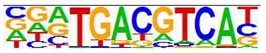
Results of c-Jun and C/EBPβ ChIP-SEQ. Human umbilical vein endothelial cells (HUVECs) were incubated for 48 h in the presence or absence of I942 or forskolin and rolipram (F/R). Following stimulation cells were fixed, chromatin was isolated and then immunoprecipitated (ChIP’d) with anti-c-Jun or anti-C/EBPβ antibodies, as described in Materials and Methods. ChIP’d DNA samples were then sequenced (ChIP-SEQ) on an Illumina GA IIx DNA sequencer. ChIP analysis of the resulting DNA sequences was then performed using the Homer (version 3.9) suite of tools [23]. The figure shows part of the Homer analysis indicating that aligned sequences from each ChIP experiment contained bone fide AP-1 [32] and C/EBP [33] consensus binding motifs, hence validating the experimental technique.
| ChIP | Treatment | Motif | Alignment | Name | p-Value |
|---|---|---|---|---|---|
| C/EBPβ | F/R |

|
RTGTTGCAA--TTNNNCAAY | CEBP/AP1 | 0.001 |
| C/EBPβ | I942 |
|
TGACGTCATGACGTCA | CREB/ATF | 0.001 |
| c-Jun | F/R |

|
RTGTTGCAA--TTNNNCAAY | CEBP/AP1 | 0.001 |
| c-Jun | I942 |

|
TGASTCATGAGTCA | Jun/AP1 | 0.001 |
