Summary
The distinct states of pluripotency in the pre- and post-implantation embryo can be captured in vitro as naive and primed pluripotent stem cell cultures, respectively. The study and application of the naive state remains hampered, particularly in humans, partially due to current culture protocols relying on extraneous undefined factors such as feeders. Here we performed a small-molecule screen to identify compounds that facilitate chemically defined establishment and maintenance of human feeder-independent naive embryonic (FINE) stem cells. The expression profile in genic and repetitive elements of FINE cells resembles the 8-cell-to-morula stage in vivo, and only differs from feeder-dependent naive cells in genes involved in cell-cell/cell-matrix interactions. FINE cells offer several technical advantages, such as increased amenability to transfection and a longer period of genomic stability, compared with feeder-dependent cells. Thus, FINE cells will serve as an accessible and useful system for scientific and translational applications of naïve pluripotent stem cells.
Keywords: endogenous retroviral element, small-molecule screens, naive human pluripotent cell state, feeder-independent and chemically defined culture, dasatinib, BCR-ABL and SRC inhibitor, AZD5438, CDK1/2/9 inhibitor
Graphical Abstract
Highlights
-
•
High-throughput screen identifies small molecules modulating human naive pluripotency
-
•
Induction and culture of human feeder-independent naive embryonic (FINE) stem cells
-
•
FINE cells are molecularly equivalent to 4iLA hESCs
-
•
FINE cells offer enhanced genomic stability and amenability to exogenous DNA uptake
In this article, Chan and colleagues develop a chemically defined culture system for the establishment and maintenance of human feeder-independent naive embryonic (FINE) stem cells. FINE cells offer several technical advantages beyond feeder independence such as enhanced genomic stability, increased amenability to transfection, and expression of 8-cell-to-morula stage markers.
Introduction
Mammalian embryonic development occurs via systematic and dynamic transitions through multiple sequential stages (Zernicka-Goetz et al., 2009). For the pluripotent cells of the epiblast, the transition from the pre-implantation to post-implantation embryo is the major landmark when fundamental molecular changes occur, distinguishing two distinct states of pluripotency. These states can now be captured in vitro as naive and primed states (Nichols and Smith, 2009), respectively, in both mouse (Brons et al., 2007, Evans and Kaufman, 1981, Martin, 1981, Tesar et al., 2007, Ying et al., 2008) and human pluripotent stem cell cultures (Chan et al., 2013, Gafni et al., 2013, Guo et al., 2017, Reubinoff et al., 2000, Takashima et al., 2014, Theunissen et al., 2014, Thomson et al., 1998, Ware et al., 2014). While the molecular machinery has been extensively studied for human primed, mouse primed, and mouse naive pluripotency states, the regulatory pathways governing the human naive state remain to be dissected. This endeavor is crucial, because: (1) naive human embryonic stem cells (hESCs) serve as a useful in vitro model of early human development, which is practically and ethically challenging to study in vivo; (2) naive cultures are more favorable than primed cultures in certain biological aspects—for example, the latter exhibits higher heterogeneity and more variability during multi-lineage differentiation (Nishizawa et al., 2016); and (3) it has been put forward that certain small molecules act differently in mouse and human pluripotent states (Ware, 2017, Weinberger et al., 2016). A major hurdle for studying the human naive state is that, unlike mouse naive and human primed states, its establishment and/or maintenance remains dependent on feeders. A defined feeder-free culture condition for the in vitro counterpart of human pre-implantation blastocyst will ease the mechanistic dissection of naive identity and facilitate the use of these cells in the clinic.
Consistent with different signaling requirements, naive cells are molecularly distinct from primed conventional human pluripotent cultures. They express naive-specific transcription factors such as KLF4, KLF5, DPPA3, and DPPA5, express higher levels of NANOG, display nuclear-specific localization of TFE3, and preferentially utilize the distal POU5F1 enhancer (Betschinger et al., 2013, Theunissen et al., 2014, Theunissen et al., 2016). These characteristics and their overall transcriptome closely resemble the in vivo intracellular matrix of human pre-implantation blastocyst (Theunissen et al., 2016). Notably, the naive and primed pluripotent states are each associated with a distinct repertoire of expressed transposons, robustly reflecting profiles of their counterparts in vivo (Göke et al., 2015, Theunissen et al., 2016). For example, primed hESCs are maintained by expression of HERVH driven by the LTR7 element (Lu et al., 2014), while naive hESCs are marked by activity of the LTR7Y elements (Göke et al., 2015, Theunissen et al., 2016) as well as expression of HERVK driven by LTR5_Hs (Grow et al., 2015, Theunissen et al., 2016). The high specificity of ERV promoters, especially throughout the course of embryonic development (Göke et al., 2015), provides a unique approach for identification of cell states beyond existing in vitro models.
In this study, we took advantage of the specific activity of LTR7Y in naive pluripotency as a tool for establishment of feeder-free naive culture conditions. By combining a sensitive stage-specific endogenous retrovirus (ERV) reporter with a high-throughput chemical screen, we identified novel molecules that we utilized to create human feeder-independent naive embryonic (FINE) stem cells.
Results
Small-Molecule Screen for Conditions Supporting Maintenance of the Human Naive Pluripotent State in the Absence of Feeders
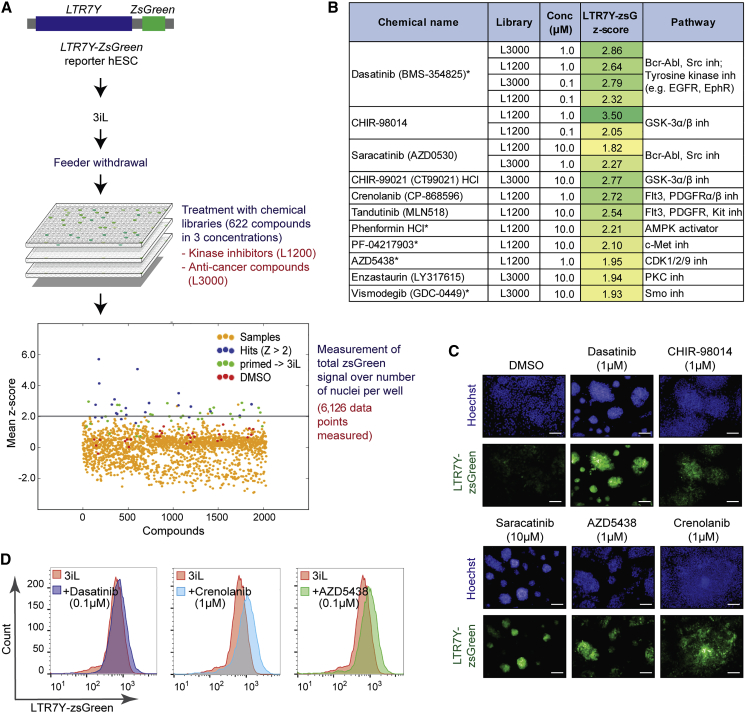
We sought a culture condition that would enable the propagation of naive hESCs without feeders through a high-throughput small-molecule screen (Figure 1A). To visualize the naive state, we developed a zsGreen reporter cell line driven by the ERV element LTR7Y, whose expression has been shown to be specific to pre-implantation blastocyst stage embryos (Göke et al., 2015). We confirmed that this reporter line is pluripotent (Figures S1A–S1C), is karyotypically normal (Figure S1D), fluoresces only in naive cells (3iL [Chan et al., 2013]) and not in primed or differentiated cells (Figures S1E and S1F), and loses naive markers and zsGreen fluorescence upon transfer to feeder-free culture (Figures S1F–S1H). To identify chemicals that can prevent collapse of naive hESCs upon feeder withdrawal, we passaged hESCs cultured in 3iL (Chan et al., 2013) onto reduced Matrigel, and after attachment added small molecules into the medium (Figure 1A). For controls, we designated wells for hESCs treated only with DMSO vehicle, hESCs cultured in mTeSR (primed, thus showing baseline fluorescence), and primed hESCs freshly transferred to 3iL (primed → 3iL; this initial conversion exhibits an increase in signal despite the absence of feeders). We screened a total of 622 compounds targeting signaling pathways governing embryonic development, cell proliferation, and cell survival. The degree of preservation of the naive state was measured through the average LTR7Y-zsGreen fluorescence intensity per cell, 4 days after feeder withdrawal. The screen was performed across three concentrations for each compound and in triplicate, summing up to 5,967 data points (Table S1).
Figure 1.
Small-Molecule Screen for Feeder-free Maintenance of Naive hESCs
(A) Schematic of high-throughput screen performed to identify compounds supporting feeder-free culture of naive hESCs. Dot plot presents mean Z scores for LTR7Y-zsGreen intensity results from the screen. The gray line indicates a cutoff of Z scores ≥2. Small molecules achieving this cutoff in at least two replicates were considered as hits (blue). Other samples (orange) and DMSO controls (red) did not pass this cutoff. A full list of scores is given in Table S1.
(B) Summary table with hits from the small-molecule screen. Asterisks denote compounds targeting pathways not previously demonstrated to play a role in establishment/maintenance of naive pluripotency.
(C) Representative images of LTR7Y-zsGreen cells after treatment with small-molecule hits. Scale bars, 50 μm.
(D) Fluorescence-activated cell sorting (FACS) quantification of LTR7Y-zsGreen signal after treatment with dasatinib, crenolanib, and AZD5438.
We first ensured the quality of the screen by certifying the absence of intra-plate layout biases (Figure S1I), proper inter-plate alignment (Figure S1J), and good correlation between replicates (Figure S1K). Z scores were then calculated, and compounds that reproducibly scored above noise (Z > 2 in at least two replicates) were regarded as hits (Figures 1B and S1L). We also detected no significant cell number bias in hit selection (Figure S1M). Finally, we manually excluded compounds that autofluoresced in the green channel as false positives. We observe that certain pathways are targeted by multiple compounds detected as hits (Figure 1B), including those previously implicated in naive pluripotency (e.g., GSK3, SRC, PDGFR) (Takashima et al., 2014, Theunissen et al., 2014). Collectively, these ascertain that we rigorously and reliably identified compounds that could retain a naive signature upon feeder withdrawal (Figure 1B). Indeed, reporter activity of hits can be validated by visual inspection (Figure 1C) as well as by flow cytometry (Figure 1D). In addition to small molecules targeting pathways implicated in naive pluripotency, our screen identified novel regulators of naive pluripotency such as the Bcr-Abl/Src inhibitors dasatinib and saracatinib, as well as the cyclin-dependent kinase inhibitor AZD5438, which warrant further investigation.
Development of a Stable Feeder-free Human Naive Pluripotent Culture Condition
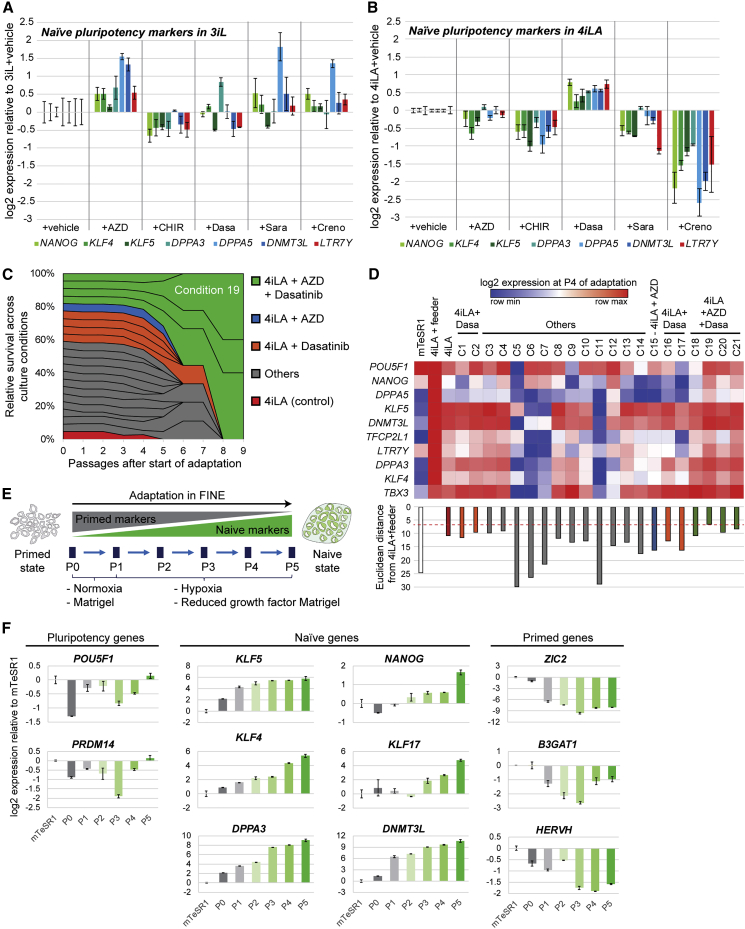
The screen provided us a list of hits that can potentially substitute for fibroblast feeders in culturing naive hESCs. To identify which of the hits might be useful for long-term culture, we first tested the effect of short-term supplementation of individual hits on naive pluripotency marker expression upon feeder withdrawal from published naive culture protocols. In 3iL (Chan et al., 2013), only AZD5438 (CDK1/2/9 inhibitor) consistently attenuated downregulation of naive pluripotency markers including LTR7Y (Figure 2A), while in 4iLA (Theunissen et al., 2014), only dasatinib (Bcr-Abl/Src kinase inhibitor) had the same effect (Figure 2B).
Figure 2.
Optimization and Establishment of FINE Culture Conditions
(A and B) Gene expression analysis for naive markers in (A) 3iL cultured cells and (B) 4iLA cultured cells supplemented with small molecules. Mean ± SD of three independent experiments. RNA was collected after 6 days (3iL) or 12 days (4iLA) in culture without feeders. AZD, AZD5438; CHIR, CHIR-99021; Dasa, dasatinib; Sara, saracatinib; Creno, crenolanib.
(C) Relative survival of hESC culture under 4iLA supplemented with different chemical combination conditions (C1–C21) over nine passages without feeders. 4iLA was included as control. When cells appear highly differentiated morphologically or when very few cells remain after passaging, the condition is dropped off; only cells cultured in C18–C21 4iLA medium supplemented with AZD5438 and dasatinib (in green) remain after nine passages. Detailed conditions are provided in Table S2.
(D) Heatmap presenting gene expression of naive pluripotency-associated markers in cells at passage 4 during adaptation to naive feeder-free conditions (C1–C21). Mean of two biological replicates is shown. Euclidean distance from 4iLA+ feeder across all the genes tested was calculated for each condition and represented as the bar chart.
(E) Schematic showing the process of adapting primed hESCs into FINE.
(F) Gene expression in hESCs throughout the course of adaptation from mTeSR1 to FINE up to passage 5. Mean ± SD of two independent experiments.
We next sought to find a condition that enables stable naive culture by adapting feeder-free primed hESCs onto media supplemented by one or more of the hits at various concentrations (Figure 2C and Table S2). We tested more combinations including AZD5438 and dasatinib because of their favorable effects on short-term feeder withdrawal (Figures 2A and 2B). From this point onward, we solely utilized 4iLA (Theunissen et al., 2014) as our basal medium, since it is the transgene-free culture condition shown to resemble the in vivo epiblast most closely at the time of the experiment (Nakamura et al., 2016). At passage 4, we collected RNA from all conditions and quantified transcripts of genes associated with naive pluripotency (Figure 2D), and found that condition 19 had the closest profile to 4iLA hESCs on feeder as measured by Euclidean distance. Despite retained expression of many naive pluripotency genes, culture in 4iLA without any feeders or supplementary molecules had very few cells surviving at passage 5. Upon culturing for eight passages, only conditions with both dasatinib and AZD5438 are still actively proliferating (Figure 2C). Since condition 19 exhibits the best survival profile while maintaining naive gene expression signature, we decided to focus on the long-term propagation of cells in this medium. Further optimization showed that WH-4-023, an Src kinase inhibitor originally present in 4iLA feeder-dependent culture (Theunissen et al., 2014), is dispensable for both adaptation and maintenance of feeder-free naive cells (Figure S2A), likely due to the presence of another Src inhibitor, dasatinib (Araujo and Logothetis, 2010), in condition 19. Thus, we excluded it from the final formulation and called this “feeder-independent naive ESCs” or FINE.
Our optimized protocol for adaptation in FINE culture conditions involves an initial conversion step from mTeSR1 medium on Matrigel to FINE under normoxia (P0), plus an additional five passages (P1–P5) under hypoxia on a reduced growth factor Matrigel substrate (Figure 2E). During this course, human pluripotency markers such as POU5F1 and PRDM14 are transiently downregulated, but return to normal levels by P5 (Figure 2F). Two trends of naive-specific marker upregulation can be observed (Figure 2F): those which are upregulated as soon as P0 and gradually increase across passages (such as KLF4 and DPPA3), and those which are not upregulated until P3 onward (such as NANOG and KLF17). Primed-specific markers are generally downregulated early on at P0–P1 (Figure 2F). Taken together, adaptation in FINE does not require feeders at any step of the process, and the naive pluripotent signature is established and stabilized by P5.
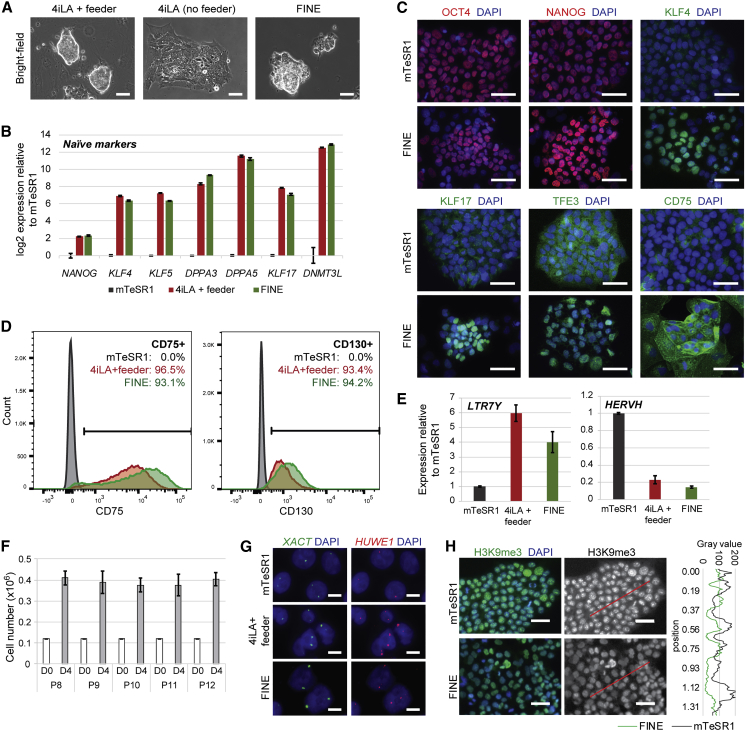
FINE Cells Exhibit Hallmarks of Naive Pluripotent Cells
Throughout propagation (P5 onward), FINE cells maintain the compact morphology characteristics of naive cells (Figure 3A). After sustained culture (>8 passages), we assessed the expression pattern of FINE compared with 4iLA on feeders and observed comparable expression levels of blastocyst markers on both transcript and protein levels (Figures 3B, 3C, and S2A), and comparable or lower expression of lineage markers (Figure S2B). Nuclear localization of naive-specific transcription factors KLF4, KLF17, and TFE3 is also observed in FINE, as in 4iLA on feeders (Figures 3C and S2A). Note that while some of these naive transcription factors exhibit heterogeneous expression, the same is observed for 4iLA on feeders (Figure S2C). Nevertheless, naive cells express naive surface markers homogeneously (Figure 3D), suggesting that all cells in culture are of naive pluripotent identity, but transcription factor levels fluctuate as observed in non-ground-state naive ESCs in mouse (Chambers et al., 2007, Hayashi et al., 2008, Niwa et al., 2009, Torres-Padilla and Chambers, 2014, van den Berg et al., 2008). Importantly, expression of stage-specific ERVs LTR7Y and HERVH in FINE mimics that of 4iLA on feeders (Figure 3E), and functional pluripotency is preserved as demonstrated by teratoma formation (Figure S2D). FINE conditions induce naivety similarly across multiple human pluripotent cell lines (Figures S3A–S3C), confirming the robustness of this culture system. To assess self-renewal capability, we quantified the cell number of FINE cells across passages and observed ∼4-fold propagation every 4 days (Figure 3F), consistent with positive staining for proliferation marker Ki67 (Figure S2E). This doubling rate (∼60 h) is comparable with that of 4iLA+ feeder (60–96 h), but is slower than primed cells in mTeSR1 (∼22 h) (Figure S2F). X chromosome status through in situ hybridization of the HUWE1 locus (Sahakyan et al., 2017) indicates XaXa for both FINE and 4iLA+ feeder, while primed cells exhibit XaXi (Figures 3G and S2G). Transcripts indicative of X chromosome activation are similarly upregulated by approximately 2-fold or more in 4iLA+ feeder and FINE versus primed cells (Figure S2H), consistent with the switch from monoallelic to biallelic expression in XaXa cells (Lin et al., 2007). We also observe that FINE cells exhibit lower levels of H3K9 trimethylation compared with primed mTeSR1 culture, typical of the higher proportion of euchromatin in ground-state naive pluripotent cells (Tosolini et al., 2018) (Figure 3H). Taken together, these results indicate that FINE is a bona fide human naive pluripotent culture system independent of feeder support.
Figure 3.
FINE Cells Display Hallmarks of Naive Pluripotency
(A) Bright-field images of hESCs cultured in 4iLA (with and without feeders) and FINE at passage 8. Scale bars, 50 μm.
(B) Expression of blastocyst-associated transcripts in hESCs cultured under mTeSR1, 4iLA+ feeder, and FINE conditions. Mean ± SD of three independent experiments.
(C) Immunofluorescence staining of pluripotency and blastocyst-associated proteins in hESCs under mTeSR1 and FINE conditions. Scale bars, 50 μm.
(D) FACS quantification of hESCs expressing naive surface markers under mTeSR1, 4iLA+ feeder, and FINE conditions.
(E) qPCR analysis of LTR7Y and HERVH transcripts in hESCs cultured under mTeSR1, 4iLA+ feeder, and FINE conditions. Mean ± SD of three independent experiments.
(F) Measurement of cell numbers cultured in FINE conditions at day 0 (D0) and 4 days post seeding (D4). Mean ± SD of three independent experiments.
(G) Representative RNA fluorescence in situ hybridization (FISH) images detecting HUWE1 (subject of X chromosome inactivation) and XACT control (escaping X chromosome inactivation) in mTeSR1, 4iLA+ feeder, and FINE cells. Scale bars, 10 μm.
(H) Immunofluorescence staining of H3K9me3 in hESCs under mTeSR1 and FINE conditions (left). Scale bars, 50 μm. Intensity of H3K9me3 was quantified through a line (red) randomly drawn across images (right).
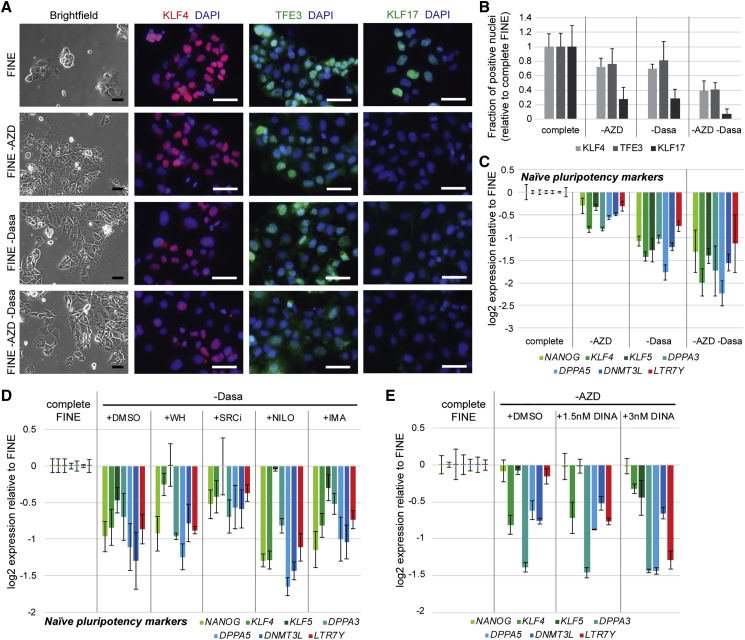
FINE Cells Are Dependent on Both Dasatinib and AZD5438
Our data showed that both AZD5438 and dasatinib are crucial for the establishment of FINE cells. Therefore, we wanted to test whether both are also important for the maintenance of naivety in FINE. Withdrawal of either or both AZD5438 and dasatinib caused dispersal of the compact morphology typical of naive cultures, indicating exit from naive pluripotency (Figure 4A). This is corroborated by the reduction in nuclear staining of naive-specific transcription factors (Figures 4A and 4B) as well as the downregulation of pluripotency and naive transcripts (Figure 4C). Dasatinib withdrawal had a more pronounced effect in loss of naivety than AZD5438 withdrawal but, more importantly, markers were lost most significantly upon withdrawal of both compounds. These results indicate that these two compounds act on distinct pathways in parallel to maintain the naive pluripotent state in the absence of feeders, and supplementation of either compound alone is insufficient to sustain long-term naivety.
Figure 4.
FINE Cells Are Dependent on Both Dasatinib and AZD5438
(A) Bright-field and immunofluorescence staining of KLF4, TFE3, and KLF17 in FINE culture after withdrawal of AZD, dasatinib (Dasa), or both for three passages. Scale bars, 50 μm.
(B) Quantification of fraction of nuclei positive for naive-associated transcription factors in hESCs cultured in FINE after withdrawal of AZD, Dasa, or both for three passages. Mean ± SD of three independent experiments.
(C) Expression of blastocyst-associated transcripts in hESCs cultured in FINE after withdrawal of AZD, Dasa, or both for two passages. Mean ± SD of three independent qPCR experiments.
(D and E) Expression of blastocyst-associated transcripts in hESCs cultured in FINE after (D) replacement of Dasa with other Src and Bcr-Abl inhibitors or (E) replacement of AZD with dinaciclib (DINA) (in H9 line) for two passages. IMA, imatinib; NILO, nilotinib. Mean ± SD of three independent qPCR experiments.
Dasatinib is a kinase inhibitor with a broad range of targets including Bcr-Abl, Src family kinases, and multiple receptor and non-receptor tyrosine kinase families (Li et al., 2010). We know that dasatinib affects Src, as its addition allowed removal of WH-4-023 from the FINE formulation (Figures 3C and S2A); however, replacement of dasatinib with other Src inhibitors such as WH-4-023 and SRCi, as well as other Bcr-Abl inhibitors such as nilotinib and imatinib (Figure 4D), failed to sustain feeder-free naive hESCs. Similarly, replacement of AZD5438 with another multi-cyclin-dependent kinase inhibitor, dinaciclib (which also inhibits CDK1/2/9, but also CDK5) did not sustain FINE cells (Figure 4E). These results establish the essential role of these two compounds in sustaining feeder-free naive culture and imply that these compounds might have other unknown targets that confer such effects.
Analysis of the Global Transcriptome of FINE Cells
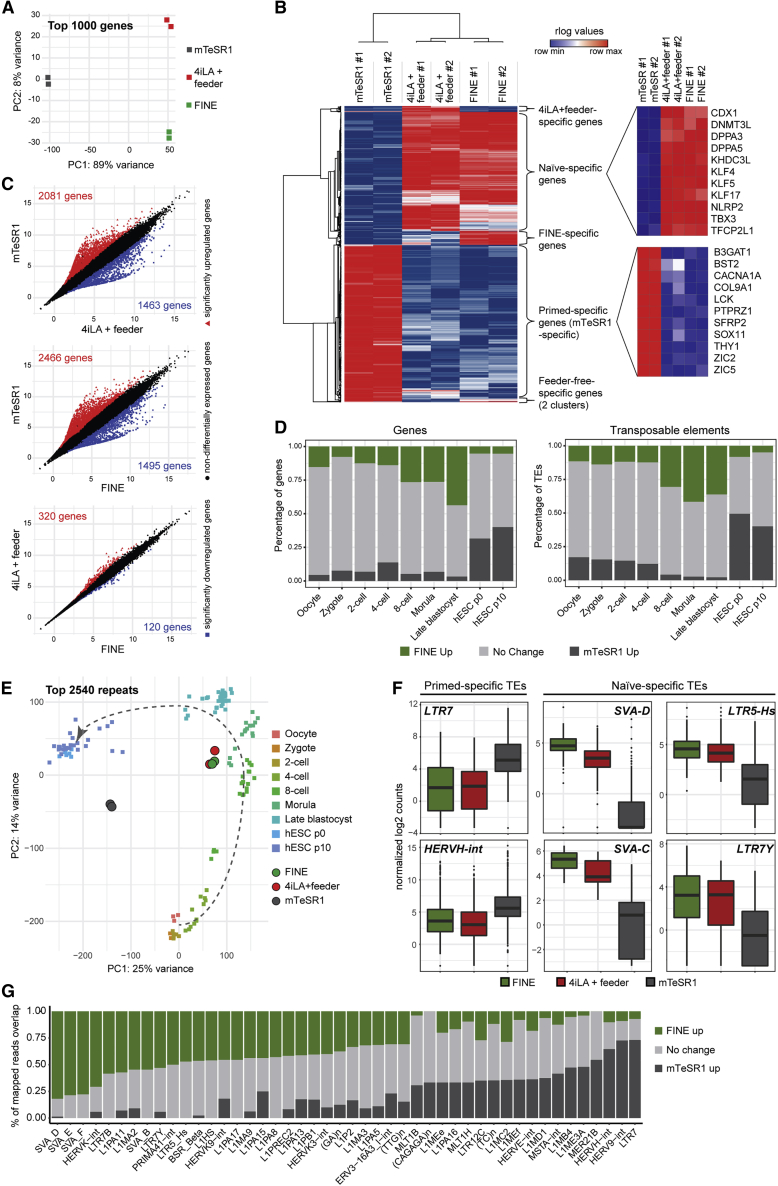
To confirm the efficacy of FINE in converting hESCs to a feeder-free naive state, we performed RNA-sequencing (RNA-seq) analysis on hESCs cultured in mTeSR1 (primed), 4iLA with feeders, and FINE. Principal component analysis and correlation confirms that FINE very closely resembles naive cells on feeders (Figures 5A and S4A). Analysis of the top 1,000 differentially expressed genes show six major clusters, with the two biggest clusters comprising genes specifically expressed in either primed (mTeSR1) or naive (4iLA+ feeder, FINE) states including well-known markers such as DNMT3L, DPPA5, KLF4, and TFCP2L1 (Chan et al., 2013, Takashima et al., 2014, Theunissen et al., 2014) (Figure 5B). In fact, differential gene expression analysis between FINE and 4iLA+ feeder only generates 440 genes (Figure 5C), most of which are involved in cell adhesion, cell-cell junctions, and extracellular matrix interactions (Figures S4B and S4C), reflective of the replacement of feeders with reduced Matrigel. Most of the FINE-enriched genes are also upregulated in feeder-free primed mTeSR1 culture (Figure S4C), suggesting that these genes play a role in adaptation to feeder-independent in vitro culture. Allocation of differentially expressed genes between FINE and mTeSR1 onto published stage-specific genes of the human developing embryo (Xue et al., 2013, Yan et al., 2013) assigns FINE closest to the late blastocyst stage in vivo and far from primed pluripotent cultures (Figures 5D [left] and S4D).
Figure 5.
Transcriptomic Profile of FINE Resembles the In Vivo Pre-implantation Blastocyst
(A) Principal component (PC) analysis based on top 1,000 differentially expressed genes between mTeSR1, 4iLA+ feeder, and FINE cultured cells.
(B) Heatmap of top 1,000 differentially expressed genes between mTeSR1, 4iLA+ feeder, and FINE cultured cells. Six main clusters were defined by dendrogram (left). Representative genes from two main clusters (naive-specific genes, primed-specific genes) are presented in smaller heatmaps (right).
(C) Scatterplots showing significantly upregulated (in red) and downregulated (in blue) genes between: mTeSR versus 4iLA+ feeder, mTeSR1 versus FINE, and 4iLA+ feeder versus FINE conditions. Genes not differentially expressed are presented in black.
(D) Correspondence between gene expression (left) or transposable element (TE) expression (right) between our naive/primed ESCs and single-cell human embryonic stages from Yan et al. (2013). For each embryonic stage, the percentage of genes/TEs with expression upregulated in FINE (green), upregulated in mTeSR1 (dark gray) or unchanged between FINE and mTESR1 (light gray) is shown.
(E) PC analysis plot based on the top 2,540 repeat elements differentially expressed across conditions. Single-cell in vivo embryonic data (Yan et al., 2013) are represented as squares, while FINE, 4iLA+ feeder, and mTeSR1 from our bulk RNA-seq data are drawn as circles.
(F) Box plots representing mean normalized expression of different TEs in mTeSR1, 4iLA+ feeder, and FINE cultured cells.
(G) Percentage of members in each TE family with expression upregulated in FINE (green), upregulated in mTeSR1 (dark gray), or unchanged between FINE and mTESR1 (light gray). TE families were ranked as specific to FINE conditions on the left and specific to mTeSR1 conditions on the right.
Expression profiles of repetitive and transposable elements have been demonstrated to be highly stage specific (Göke et al., 2015), and can be used as a sensitive barometer for matching pluripotent cultures with stages of in vivo early human development (Theunissen et al., 2016). Analysis of this “transposcriptome” matched FINE cells with the 8-cell-to-blastocyst stages of the human embryo (Figures 5D [right] and 5E), with FINE cells very closely resembling hESCs cultured in 4iLA+ feeder (Figures 5E, 5F, and S4E). Transposable elements upregulated in FINE versus primed cells include known naive-specific families such as LTR5-Hs, LTR7Y, and HERVK (Göke et al., 2015, Grow et al., 2015, Theunissen et al., 2016) (Figures 5F and 5G). Overall, FINE cells are transcriptionally equivalent to naive pluripotent culture with feeders, and closely resemble the in vivo pre-implantation blastocyst.
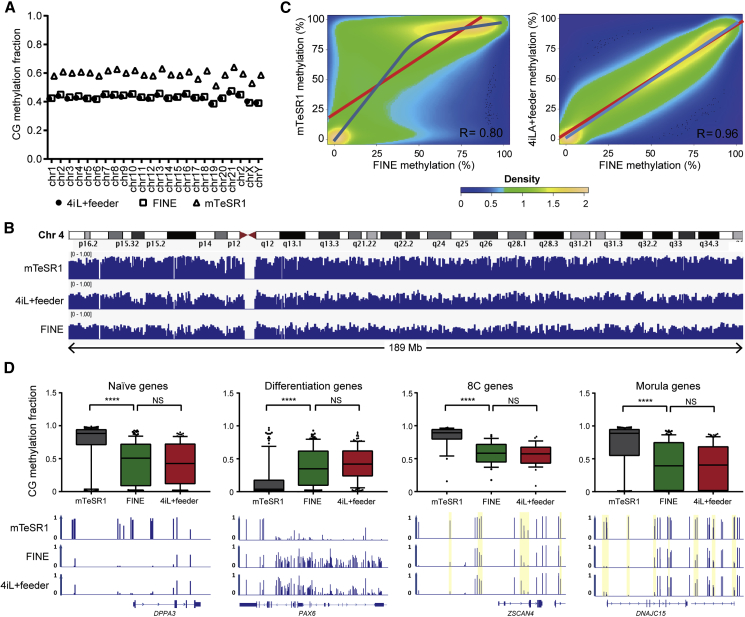
Analysis of Global DNA Methylation in FINE Cells
To examine the chromatin status of FINE cells, we profiled global DNA methylation status in FINE cells compared with mTeSR1 and 4iLA+ feeder cells. Across all chromosomes, the percentage of methylated CG sites is equivalently lower in cells cultured in both naive conditions compared with primed hESCs (Figures 6A and 6B), consistent with previous reports of global DNA hypomethylation in the naive pluripotent state (Takashima et al., 2014, Theunissen et al., 2016). In fact, the methylated regions for FINE and 4iLA+ feeder are very highly correlated (Figures 6B and 6C), corroborating that these naive pluripotent states are equivalent. Consistent with transcript expression patterns, DNA at naive marker loci, as well as at 8C and morula-associated gene loci, are less methylated in FINE and 4iLA+ feeders, while differentiation genes display higher DNA methylation (Figure 6D). All in all, global DNA methylation profiling supports that FINE represents a feeder-free equivalent of 4iLA-cultured naive hESCs.
Figure 6.
Global DNA Methylation Profile of FINE Confirms Equivalence to Feeder-Dependent Naive Pluripotent hESCs
(A) Per-chromosome comparison of CG methylation fraction between mTeSR1, 4iLA+ feeder, and FINE conditions.
(B) Relative methylation tracks of chromosome 4 under mTeSR1, 4iLA+ feeder, and FINE conditions.
(C) Correlation plot of methylated sites in FINE versus either mTeSR1 or 4iLA+ feeder. Red line represents fit based on linear regression modeling (off-center best fit indicates lower correlation); blue line is based on LOESS weighted regression modeling (curved best-fit line indicates non-linear correlation).
(D) Box plots (top) for CG methylation fraction at select loci representing naive-, differentiation-, 8C-, and morula-associated genes, as well as relative methylation tracks of one representative gene per group (bottom). Differential peaks are highlighted in yellow for ZSCAN4 and DNAJC15.
Advantages and Applications of FINE Cells
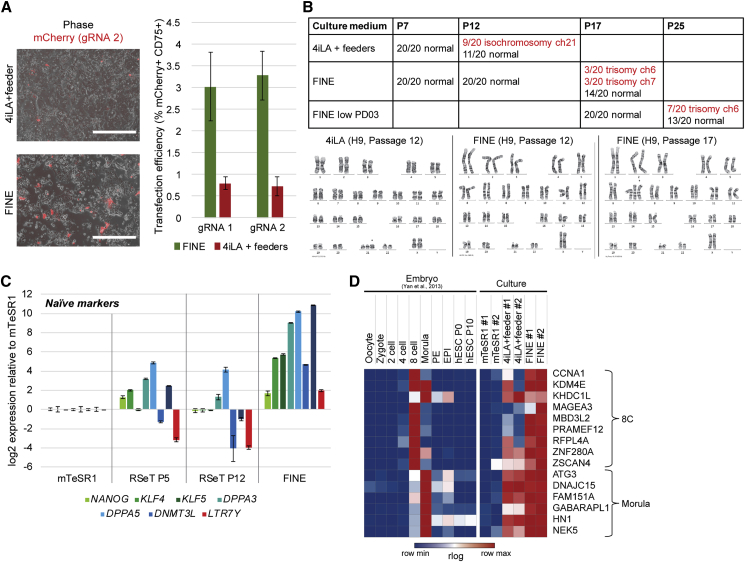
To test the utility of FINE cells for applications such as genetic targeting, we compared the amenability of naive cells to DNA delivery via transfection. Using an mCherry-expressing plasmid to report transfection efficiency and by co-staining with CD75 to exclude feeders from quantification, we observed more than 5-fold double-positive cells in FINE than in 4iLA+ feeder cells (Figures 7A and S5A). Moreover, we tested the ease of genome editing under FINE using the CRISPR/Cas9 system. Using guide RNAs targeting human-specific sequences, we observed that hESCs had no significant difference in terms of gene-editing efficiency between FINE and 4iLA+ feeder conditions (independent of transfection efficiency, as only positively transfected cells were utilized) (Figure S5B). Thus, combined with the general simplicity of handling associated with the absence of feeders, these results indicate that FINE culture allows for easier genetic targeting of naive hESCs.
Figure 7.
FINE Cells Offer Advantages over Other Naive Culture Conditions
(A) Representative images (left) and FACS quantification (right) of cells in FINE and 4iLA+ feeder culture conditions after transfection with mCherry-containing plasmids gRNA 1 (targeting EGFR) and gRNA 2 (targeting STAG2). Quantification was performed after staining with an anti-CD75 antibody to account for feeders; mean ± SD of two independent experiments. Scale bar, 400 μm.
(B) Summary of cytogenetic analysis of H9 cells (top) under various naive culture conditions (rows) and passage numbers (columns). Representative karyotypes at various passage numbers in FINE (bottom).
(C) qPCR analysis of naive-associated transcripts in H1 hESCs cultured under mTeSR1, RSeT, and FINE conditions. Mean ± SD of three independent experiments.
(D) Heatmap showing rlog values for expression of 8-cell- and morula-stage-associated genes in mTeSR1, 4iL+ feeder, and FINE cultures based on RNA-seq.
One of the main disadvantages of current human naive culture conditions is its inherent genomic instability (Theunissen et al., 2014). FINE cells are karyotypically normal up to passage 12, indicating that FINE cells are not an artifact of spontaneous genetic abnormalities (Figure 7B). Importantly, comparative cytogenetic analysis across various passage numbers indicate that FINE cells acquire chromosomal abnormalities at a slower rate than 4iLA+ feeder cells (Figure 7B), albeit having similar proliferation kinetics (Figure S2F). To further improve this, we tried lowering the concentration of PD0325901 in FINE, following a recent study that demonstrated enhanced genetic stability upon lower dosage of MEK inhibition (Di Stefano et al., 2018). While this further delayed the acquisition of chromosomal abnormalities (Figure 7B), it had a slight adverse effect on naive pluripotent marker expression (Figure S5C). Conversely, extended culture in FINE (P24) does not repress (but actually even slightly increases) naive marker expression, despite chromosomal defects (Figure S5D).
We also compared how FINE fares against RSeT, a commercially available feeder-free medium for naive hESCs, based on Gafni et al. (2013). We find that upregulation of naive markers is greater and more consistent in FINE (Figure 7C), including markers that have been reported to be upregulated by RSeT culture. Finally, we also observe that markers associated with earlier stages of development (such as the 8-cell stage) are slightly enhanced in FINE compared with 4iLA on feeder (Figures 7D and S5E). Taken together, FINE improves on existing naive culture conditions by offering robust naive marker expression, including 8-cell stage-specific transcripts, under feeder-free conditions, while improving genomic stability and amenability to gene-editing techniques.
Discussion
ERVs as Molecular Landmarks for Cellular States
Here, we exploited the stage-specific transcription of ERVs during embryogenesis to generate an ERV-based LTR7Y fluorescent reporter, and demonstrated its utility in an unbiased chemical screen that led us to establishment of a feeder-free naive medium composition. Our results and research in mouse ESCs (Macfarlan et al., 2012) indicate that with the help of accurate and sensitive reporters such as ERVs, it is possible to isolate a cell state beyond existing in vitro models. Application of a similar strategy can be used for generation of cellular models that correspond to cells from stages of development other than the blastocyst, and enable further understanding of the requirements for establishment of cellular potency and initiation of cell-fate decisions, which is still largely inaccessible to research.
Applications of Feeder-free Naive hESCs
Until now a number of protocols using various cocktails of molecules has been reported to induce naive states mimicking in vitro human pre-implantation epiblast (Chan et al., 2013, Gafni et al., 2013, Takashima et al., 2014, Theunissen et al., 2014, Ware et al., 2014). In these studies, establishment of human naive medium composition was guided by previous knowledge obtained from the mouse. However, species-specific differences exist between mouse and human pluripotency. For example, GSK3 inhibitor is commonly used in naive cocktails, and acts in mouse ESCs by elevating levels of Esrrb (Martello et al., 2012). In contrast, ESRRB is not expressed in human pluripotent states (Weinberger et al., 2016) either in vivo (blastocyst's inner cell mass) or in vitro (primed and naive hESCs), suggesting that GSK3 inhibition might act via a different mechanism in human naive pluripotent culture. In addition, naive hESCs hitherto have been dependent on feeders, a fact that introduces a non-defined component and hampers both their acceptance in clinical use and the application of certain technical approaches for dissection of mechanisms behind the state. So far, there are only two feeder-free alternatives for naive culture of hESCs: the first is RSeT medium (based on Gafni et al., 2013), which we and others have shown to have divergent transcriptional and epigenetic profiles from best-in-class naive culture systems (Barakat et al., 2018, Nakamura et al., 2016) (Figure 7C); the second is the protocol from Smith and colleagues (Guo et al., 2017), whose caveats include the requirement for HDAC inhibitors, which are known to increase susceptibility to genomic instability (Eot-Houllier et al., 2009), and the multi-step derivation process that still undergoes temporary culture on feeders for stabilization. Here, we developed a simple feeder-independent system called FINE that can be used for both the establishment and sustenance of the human naive state. Conversion to naive cells in FINE is a one-step process that does not require the use of non-defined components. Therefore, FINE provides a purely chemically defined xeno-free platform for further dissection of the mechanisms controlling human early development. This is especially useful in dissecting the role of the various small molecules that define human naive pluripotent culture. Absence of feeders also allows for unbiased high-throughput screens for identification of novel contributors to the naive state without the complication of secondary phenotypes from extraneous supporting cells not normally found in embryonic development in vivo. FINE also enables easy genetic targeting of naive hESCs, not only through the ease of handling due to its feeder-free nature (e.g., removing the requirement for antibiotic-resistant feeders for selection), but also because of its inherent amenability to such techniques (Figures 7A, S5A, and S5B). We also observe that FINE cells acquire chromosomal abnormalities slower (Figure 7B), suggesting that our system also improves genetic stability compared with its feeder-dependent counterparts. Thus, FINE culture has the potential to be the go-to system for establishment, propagation, and examination of human naive pluripotent cells.
Mechanism of Action of Effective Compounds in FINE
The unique ability of FINE to support naive hESCs in the absence of feeders is endowed by the synergistic action of two compounds, AZD5438 and dasatinib. Dasatinib is a broad kinase inhibitor affecting multiple receptor and non-receptor tyrosine kinase families (Li et al., 2010). One of its known targets that play a role in naive pluripotency is Src, but it must target other additional pathways because other Src inhibitors are unable to sustain feeder-free naive hESCs (Figure 4D). On the other hand, AZD5438 inhibits cyclin-dependent kinases 1 and 2, which largely act in checkpoints of the S and G2 phases of the cell cycle. We have demonstrated before that prolonging these phases of the cell cycle restricts exit from pluripotency in primed hESCs (Gonzales et al., 2015), and AZD5438 might act in this manner to maintain pluripotency in the absence of feeders. Yet, like dasatinib, replacement of AZD5438 with another CDK1/2/9 inhibitor was not sufficient to sustain FINE cells. Thus, while beyond the scope of this paper, it is tempting to speculate that these compounds, together or alone, affect unprecedented pathways to induce and maintain feeder-free naivety. Furthermore, several of these compounds' known target pathways have not been studied in the context of naive pluripotency, and, as such, dissecting the mechanisms of these compounds will be an exciting avenue to pursue in the future. Finally, given that these compounds were discovered in an unbiased screen, it is possible that the combination of AZD5438 and dasatinib, and perhaps other hits from our screen, may be applicable to endow feeder independence in other culture systems beyond naive hESCs.
In conclusion, we first identified novel molecules that facilitate feeder independence of naive hESC culture, which could be useful to guide studies in understanding how fibroblast feeders provide an artificial niche for stem cell culture in vitro. Second and more importantly, through rigorous optimization, we developed a simple chemically defined xeno-free method of establishing and maintaining naive hESCs called FINE. This platform offers technical advantages for the mechanistic dissection of naive identity and will serve as a useful foundation for translational applications of naive pluripotent stem cells.
Experimental Procedures
All animal experiments were approved by the A∗STAR Institutional Animal Care and Use Committee (IACUC) following the National Advisory Committee for Laboratory Animal Research (NACLAR) Guidelines. All animals were kept in pathogen-free conditions in the AAALAC-accredited A∗STAR animal facility.
Human Primed and Naive Cell Cultures
H1 (WA-01) line was used for all experiments unless specified otherwise. Other lines used are HES3 (ES-03), H9 (WA-09), and induced pluripotent stem (GM23338) cells. Primed hESCs were propagated in mTeSR1 (STEMCELL Technologies). Cells were cultured on 30× diluted Matrigel matrix-coated (Corning) dishes under normoxia (37°C, 21% O2, 5% CO2). Culture medium was refreshed daily. Cells were subcultured using 1 mg/mL dispase in DMEM/F12 (STEMCELL) every 3–6 days according to manufacturer's protocol. 3iL cultured cells were propagated as previously described (Chan et al., 2013). 4iLA+ feeder cells were cultured as previously described (Theunissen et al., 2014). Cells were cultured in hypoxic conditions (5% O2, 5% CO2). Medium was refreshed daily. Cells were subcultured using TrypLE (Life Technologies) every 4–7 days. H1 mTeSR cells were adapted to RSeT feeder-free culture conditions following the manufacturer's protocol (STEMCELL). Cells were cultured in hypoxic conditions (5% O2, 5% CO2).
Conversion of Primed hESCs to Naive hESCs with FINE Culture
Primed cells were seeded on 30× diluted Matrigel at a passage ratio of 1:6. Cells were seeded in clumps and kept in mTeSR1 culture for 48 h. For conversion to naive cell state, we removed mTeSR1 medium and added FINE culture medium, which consists of a basal medium (1:1 ratio of F12 DMEM and Neurobasal [Gibco] medium, 1× N2 supplement [Gibco], 1× B2 supplement [Gibco], 1× L-glutamine (Gibco), 1× non-essential amino acids [Gibco], 0.1 mM β-mercaptoethanol [Sigma], and 62.5 ng/mL BSA [Sigma]) supplemented with 0.1 μM dasatinib (Selleckchem), 0.1 μM AZD5438 (Tocris), 0.1 μM SB590885 (Sigma), 1 μM PD0325901 (Sigma), 10 μM Y-27632 (STEMCELL), 20 ng/mL human recombinant leukemia inhibitory factor (Peprotech), 20 ng/mL activin A (STEMCELL), and 8 ng/mL basic fibroblast growth factor (Gibco). Cells were incubated under normoxic conditions (21% O2, 5% CO2) for 4–5 days. FINE culture medium was replenished daily. At the end of conversion, cells were passaged as single cells using TrypLE (Gibco) solution on Reduce Growth Factor Matrigel-coated (Corning) plates (dishes were coated for at least 1 h before use). In brief, cells were washed with 1× PBS, and 500 μL of TrypLE was added to each 3.5-cm well (6-well plate, Falcon) of hESCs. Cells were incubated at 37°C for 1–2 min. When cells started to detach from each other and remained adherent to the plate, TrypLE was aspirated thoroughly and washed with 1× PBS. After adding 1 mL of FINE medium, cells were gently detached using a cell scraper and clumps dissociated to single cells with a 1-mL pipette. Cells were seeded at a high ratio of 1:2 in coated plates and transferred to a hypoxia (5% O2, 5% CO2) incubator for subsequent culture. Medium was refreshed daily. FINE culture cells were subsequently passaged at ratios of 1:2 to 1:4. For most cell lines, differentiated cells were observed in the first 2–3 passages and gradually decreased over subsequent passages. For the experiments described, FINE cells were cultured in medium for at least five passages before use, unless otherwise described.
FINE low PD03 cells were adapted from FINE conditions at passage 12. PD0325901 (Sigma) concentration was reduced from 1 μM to 0.3 μM.
Reporter Line Generation
LTR7Y element (chr17:32,515,593-32,516,013, hg19) was cloned into modified pLVTH-zsGreen plasmid (Addgene #12262). LTR7Y element was inserted between the PacI and SalI cloning sites. H1 hESCs were seeded at clonal density and transduced with lentivirus in the presence of 4 μg/mL Polybrene (Sigma). Cells were reseeded as single cells for generation of clonal lines for further study.
High-Throughput Small-Molecule Screen
Three thousand cells cultured in 3iL were seeded per well into 384-well plates (Greiner) coated with 30× growth factor reduced Matrigel (Corning) in 45 μL of medium. Four hours after seeding, cells were treated with anti-cancer and anti-kinase libraries (Selleckchem; kinase inhibitor screening library: customized collection of 273 kinase inhibitors; anti-cancer compound library: customized collection of 349 bioactive compounds). Small molecules were used at three different concentrations: 100 nM, 1 μM, and 10 μM. Forty-eight hours after the treatment, culture medium was renewed concomitant with a second round of treatment. Forty-eight hours after the second round of treatment, cells were fixed with 4% formaldehyde (Sigma) and stained with Hoechst 33342 dye (1:4,000, Invitrogen). Images were taken using an Opera Phenix High-Content Screening System (PerkinElmer) at 20× magnification. Images were processed and fluorescence signal was quantified using a Columbus Image Data Storage and Analysis System (PerkinElmer). Screen analyses were carried out using Screensifter software (Kumar et al., 2013). Z score for zsGreen fluorescence was calculated using formula z = (X − μ)/s.d., where μ is the mean, s.d. the standard deviation of the whole population, and X the integrated intensity of zsGreen divided by total number of cells.
850K DNA Methylation Profiling
Genomic DNA was isolated by a DNeasy Blood & Tissue Kit (Qiagen) and processed using a Zymo EZ DNA Methylation kit (Zymo Research, CA, USA) following the manufacturer's recommendations for bisulfide conversion. An Infinium MethylationEPIC BeadChip (Illumina) was used to interrogate the genome-wide methylation profile following the Infinium HD methylation assay protocol.
Author Contributions
I.S., K.A.U.G., Y.-S.C., and H.-H.N. designed the project. I.S. and K.A.U.G. conducted most of the experiments, analyzed the results, and wrote the manuscript. E.C and J.G. conducted bioinformatics analysis. M.N.B.R., B.P.G.L., and C.P.T. performed experiments and analyzed data. H.L. performed methylation profiling. C.K.W. and G.I.R. performed FISH.
Acknowledgments
This research is supported by the Agency for Science, Technology and Research, Singapore and the Singapore Ministry of Health's National Medical Research Council under the Open Fund Individual Research Grant NMRC/OFIRG/0045/2017-00. I.S. was supported by the Singapore International Graduate Award from A∗STAR. Work in the G.I.R. lab is supported by a National Research Foundation Fellowship to G.I.R. (NRFI05-2019-0008). The authors would like to thank Singapore Center for High Throughput Phenomics at GIS, G. Periyasamy, M. Thangavelu, and also Y.S. Lim and S. Wang for assistance with experimental work. We also thank L. Yang for help with probes design for FISH and comments on the text, and A. Malinowski for comments on the manuscript.
Published: September 12, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.08.005.
Contributor Information
Huck-Hui Ng, Email: nghh@gis.a-star.edu.sg.
Yun-Shen Chan, Email: chanysw@gis.a-star.edu.sg.
Accession Numbers
RNA-seq data have been deposited in the Gene Expression Omnibus under accession number GEO: E-MTAB-8216.
Supplemental Information
References
- Araujo J., Logothetis C. Dasatinib: a potent SRC inhibitor in clinical development for the treatment of solid tumors. Cancer Treat. Rev. 2010;36:492–500. doi: 10.1016/j.ctrv.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat T.S., Halbritter F., Zhang M., Rendeiro A.F., Perenthaler E., Bock C., Chambers I. Functional dissection of the enhancer repertoire in human embryonic stem cells. Cell Stem Cell. 2018;23:276–288.e8. doi: 10.1016/j.stem.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Nichols J., Dietmann S., Corrin P.D., Paddison P.J., Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chan Y.S., Göke J., Ng J.H., Lu X., Gonzales K.A., Tan C.P., Tng W.Q., Hong Z.Z., Lim Y.S., Ng H.H. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Di Stefano B., Ueda M., Sabri S., Brumbaugh J., Huebner A.J., Sahakyan A., Clement K., Clowers K.J., Erickson A.R., Shioda K. Reduced MEK inhibition preserves genomic stability in naive human embryonic stem cells. Nat. Methods. 2018;15:732–740. doi: 10.1038/s41592-018-0104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eot-Houllier G., Fulcrand G., Magnaghi-Jaulin L., Jaulin C. Histone deacetylase inhibitors and genomic instability. Cancer Lett. 2009;274:169–176. doi: 10.1016/j.canlet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Göke J., Lu X., Chan Y.S., Ng H.H., Ly L.H., Sachs F., Szczerbinska I. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16:135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Gonzales K.A., Liang H., Lim Y.S., Chan Y.S., Yeo J.C., Tan C.P., Gao B., Le B., Tan Z.Y., Low K.Y. Deterministic restriction on pluripotent state dissolution by cell-cycle pathways. Cell. 2015;162:564–579. doi: 10.1016/j.cell.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Grow E.J., Flynn R.A., Chavez S.L., Bayless N.L., Wossidlo M., Wesche D.J., Martin L., Ware C.B., Blish C.A., Chang H.Y. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., von Meyenn F., Rostovskaya M., Clarke J., Dietmann S., Baker D., Sahakyan A., Myers S., Bertone P., Reik W. Epigenetic resetting of human pluripotency. Development. 2017;144:2748–2763. doi: 10.1242/dev.146811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Lopes S.M., Tang F., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Goh G., Wongphayak S., Moreau D., Bard F. ScreenSifter: analysis and visualization of RNAi screening data. BMC Bioinformatics. 2013;14:290. doi: 10.1186/1471-2105-14-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Rix U., Fang B., Bai Y., Edwards A., Colinge J., Bennett K.L., Gao J., Song L., Eschrich S. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat. Chem. Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Gupta V., Vermilyea M.D., Falciani F., Lee J.T., O'Neill L.P., Turner B.M. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Sachs F., Ramsay L., Jacques P.E., Göke J., Bourque G., Ng H.H. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol. 2014;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Gottgens B., Niwa H., Smith A. Esrrb is a pivotal target of the gsk3/tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Okamoto I., Sasaki K., Yabuta Y., Iwatani C., Tsuchiya H., Seita Y., Nakamura S., Yamamoto T., Saitou M. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016;537:57–62. doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nishizawa M., Chonabayashi K., Nomura M., Tanaka A., Nakamura M., Inagaki A., Nishikawa M., Takei I., Oishi A., Tanabe K. Epigenetic variation between human induced pluripotent stem cell lines is an indicator of differentiation capacity. Cell Stem Cell. 2016;19:341–354. doi: 10.1016/j.stem.2016.06.019. [DOI] [PubMed] [Google Scholar]
- Niwa H., Ogawa K., Shimosato D., Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Reubinoff B.E., Pera M.F., Fong C.Y., Trounson A., Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat. Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T.W., Kuoy E., Langerman J., Clark A.T., Jaenisch R. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2017;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen T.W., Friedli M., He Y., Planet E., O'Neil R.C., Markoulaki S., Pontis J., Wang H., Iouranova A., Imbeault M. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla M.E., Chambers I. Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- Tosolini M., Brochard V., Adenot P., Chebrout M., Grillo G., Navia V., Beaujean N., Francastel C., Bonnet-Garnier A., Jouneau A. Contrasting epigenetic states of heterochromatin in the different types of mouse pluripotent stem cells. Sci. Rep. 2018;8:5776. doi: 10.1038/s41598-018-23822-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg D.L., Zhang W., Yates A., Engelen E., Takacs K., Bezstarosti K., Demmers J., Chambers I., Poot R.A. Estrogen-related receptor beta interacts with Oct4 to positively regulate Nanog gene expression. Mol. Cell. Biol. 2008;28:5986–5995. doi: 10.1128/MCB.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware C.B. Concise review: lessons from naive human pluripotent cells. Stem Cells. 2017;35:35–41. doi: 10.1002/stem.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware C.B., Nelson A.M., Mecham B., Hesson J., Zhou W., Jonlin E.C., Jimenez-Caliani A.J., Deng X., Cavanaugh C., Cook S. Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L., Ayyash M., Novershtern N., Hanna J.H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Xue Z., Huang K., Cai C., Cai L., Jiang C.Y., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y.E. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M., Morris S.A., Bruce A.W. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat. Rev. Genet. 2009;10:467–477. doi: 10.1038/nrg2564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.