Figure 7.
FINE Cells Offer Advantages over Other Naive Culture Conditions
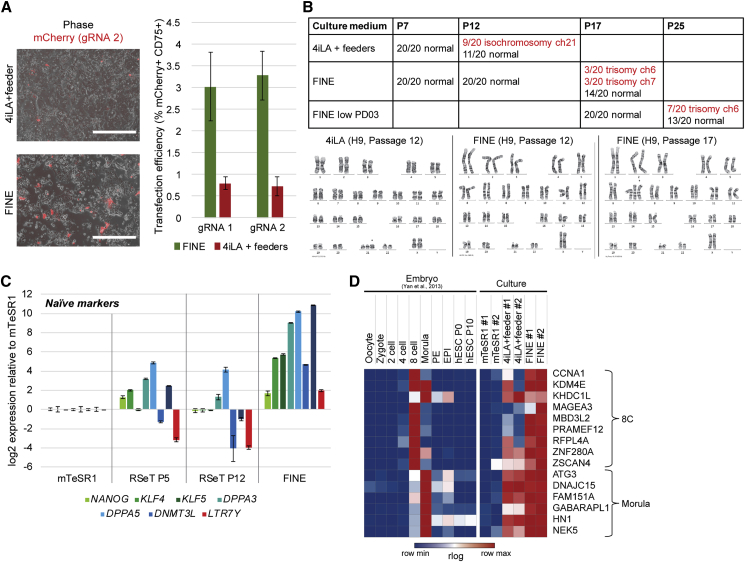
(A) Representative images (left) and FACS quantification (right) of cells in FINE and 4iLA+ feeder culture conditions after transfection with mCherry-containing plasmids gRNA 1 (targeting EGFR) and gRNA 2 (targeting STAG2). Quantification was performed after staining with an anti-CD75 antibody to account for feeders; mean ± SD of two independent experiments. Scale bar, 400 μm.
(B) Summary of cytogenetic analysis of H9 cells (top) under various naive culture conditions (rows) and passage numbers (columns). Representative karyotypes at various passage numbers in FINE (bottom).
(C) qPCR analysis of naive-associated transcripts in H1 hESCs cultured under mTeSR1, RSeT, and FINE conditions. Mean ± SD of three independent experiments.
(D) Heatmap showing rlog values for expression of 8-cell- and morula-stage-associated genes in mTeSR1, 4iL+ feeder, and FINE cultures based on RNA-seq.