Figure 4.
Properties and Molecular Signatures of Lgr5+ Cells in E14.5 Knee Joint Interzone Revealed by Single-Cell Transcriptome
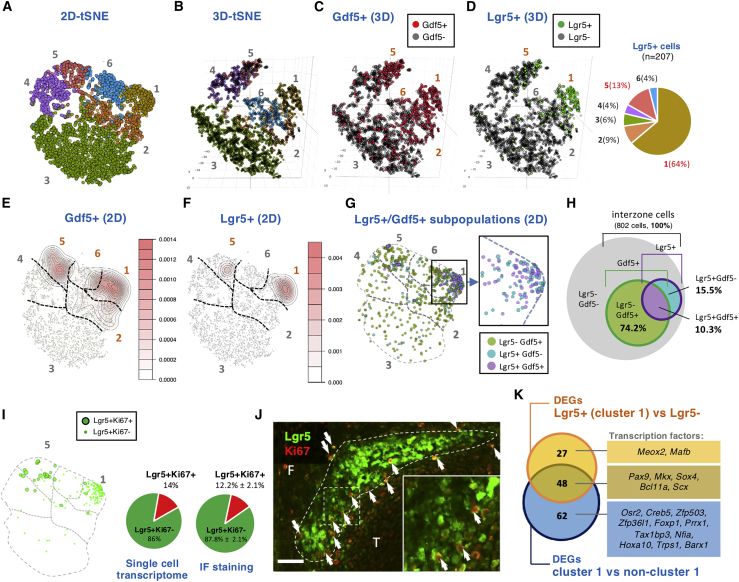
(A and B) Gene expression from 5,460 cells was analyzed using tSNE plots in 2D (A) or 3D (B), with six major clusters identified.
(C and D) 3D view for the distribution of Gdf5+ (C) and Lgr5+ (D) cells. The relative distribution of Lgr5 expression in the different clusters is presented as a pie chart in (D).
(E and F) Contour plots illustrating the density of cells expressing (F) Lgr5 shown in (B), and (E) Gdf5 shown in (C), with the corresponding colored density chart.
(G) 2D-tSNE showing the distribution of Lgr5+ among Gdf5+ cells. Higher magnification of the boxed area in cluster 1 is shown on the right panel, highlighting the relative Lgr5+/Gdf5− (cyan) and Lgr5+/Gdf5+ (purple) cells.
(H) Venn diagram illustrating the distribution and percentages of Lgr5−/Gdf5+ (green), Lgr5+/Gdf5− (cyan), and Lgr5+/Gdf5+ (purple) subpopulation of cells.
(I) 2D-tSNE plot highlighting the distribution of Lgr5+/Ki67+ in cluster 5, relative to the dominance of Lgr5+/Ki67− cells in cluster 1, and the percentage of Lgr5+/Ki67+ shown in a pie chart.
(J) Double immunofluorescence detection of Lgr5-GFP- and Ki67-expressing cells in an E14.5 knee joint. The peripheral of the Lgr5 cell population is marked with a dotted line, and arrows indicate the double-positive cells with the boxed region shown in higher magnification. The relative percentage of Lgr5+/Ki67+, determined from three embryos, is also shown in a pie chart. F, femur; T, tibia. Scale bars, 20 μm.
(K) Venn diagram showing the DEGs of cluster 1 versus non-cluster 1 (n = 110, blue circle) and DEGs Lgr5+ versus Lgr5− cells (n = 75, yellow circle). There are 48 common DEGs identified with these two gene lists. Transcription factors in each sector are shown.