Figure 5.
Lineage Trajectories of Knee Joint
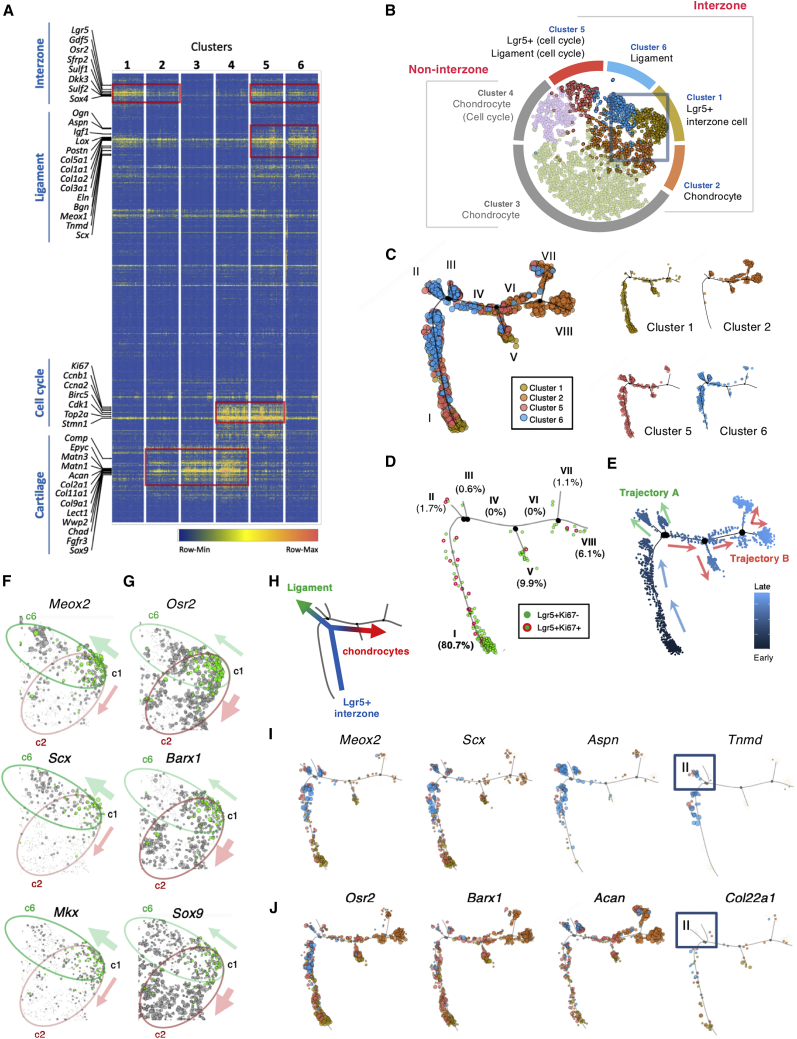
(A) Heatmap showing the clustering of the top 5% dispersed genes (genome-wide), from the profiles of 200 cells randomly selected from each cluster as a representative. Gene clusters related to joint development, ligament, cell cycle, and cartilage were identified.
(B) Summary diagram of the property/signature of each cluster, with clusters 1, 2, 5, and 6 defined as interzone cells.
(C) Pseudo-timeline analysis of cells from clusters 1, 2, 5, and 6 identified eight cell states, and the distribution of cells from individual cluster and a combined map along the pseudo-timeline is shown.
(D) Distribution of all Lgr5+ cells along the pseudo-timeline, and relative percentages distributed in each of the cell state indicated, together with proliferative Lgr5+/Ki67+ cells.
(E–G) The pseudo-timeline (E) predicted a gradient of “early” (dark blue) to late (light blue) cells, with a major divergence to two trajectories A and B, with arrows indicating the predicted direction. Expression distribution of ligament (F) and chondrocyte (G) genes are mapped onto the 2D tSNE. The size of each circle is a relative reflection of the gene expression level; Lgr5+ cells are indicated as green.
(H) Illustration showing the major lineage divergence of Lgr5+ interzone cells.
(I and J) Distribution of cells expressing ligament (I) and cartilage (J) genes along the pseudo-timeline. The boxes highlight a specific comparison of cells in state II expressing a mature ligament marker Tnmd but not Col22a1.