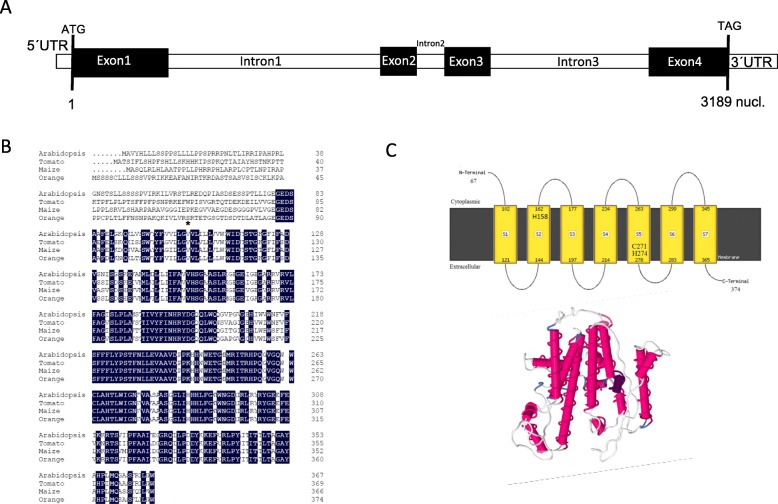
Fig. 5.
Sweet orange Z-ISO gene and protein structures. a Structure of sweet orange Z-ISO gene with four exons (black) and three introns (white). b Alignment of Z-ISO proteins from sweet orange (KDO50569.1), Arabidopsis (NP563879.1), maize (NP_001132720.1) and tomato (XP_004252966.1). Numbers on the left denote the number of amino acid residues and identical for all sequences in a given position are in black background. The end of predicted a plastid transit peptide marked with an asterisk. c Sweet orange Z-ISO topology predicted by MEMSAT3 and tridimensional modelling of Z-ISO (PPMserver) by using iTASSER. Transmembrane α-helix S1 to S7 are represented in yellow and pink in the 2D and 3D-models, respectively