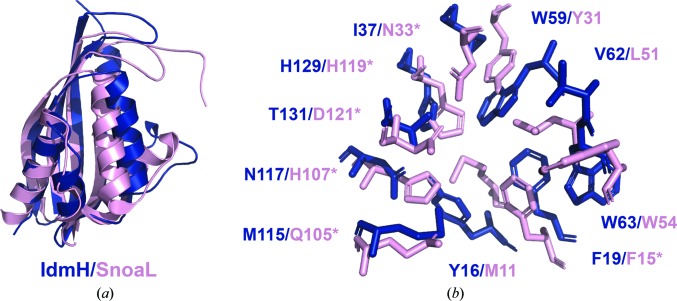
Figure 4.
Comparison of IdmH (blue) with its sequence and structural homologue SnoaL (pink). (a) IdmH chain A was aligned with SnoaL (PDB entry 1sjw) chain A using PyMOL. (b) Active-site residues in SnoaL (pink) compared with their counterparts in IdmH (blue). While some residues appear to be conserved between the two proteins (His129/His119, Trp63/Trp54 and Phe19/Phe19), the charged essential catalytic residues His107 and Asp121 are unique to SnoaL. Another difference between the two active sites arises from different choices and positioning of aromatic residues (Trp59/Tyr31 and Trp63/Trp54), while Tyr16 appears to be completely unique to IdmH. Asterisks denote SnoaL catalytic residues (Sultana et al., 2004 ▸).