Table 2. β2AR ligands used in the exchange experiments with their MoAs, molecular weights (MW) and affinity (K i) values.
Data are from the ChEMBL database (ChEMBL_23; Gaulton et al., 2012 ▸). Chemical structures of the ligands used for the other receptors in this study are shown in Supplementary Fig. S4.
| Ligand | Chemical structure | MoA | ChEMBL ID | MW (Da) | K i (nM) | Reference |
|---|---|---|---|---|---|---|
| ICI-118,551 |
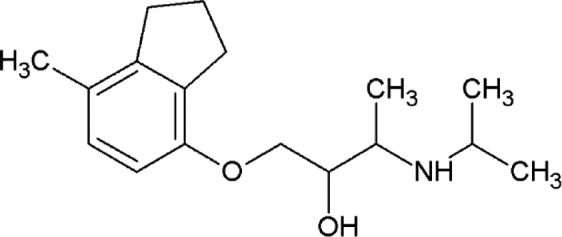
|
Inverse agonist | CHEMBL513389 | 277.4 | 0.13 | Dolušić et al. (2011 ▸) |
| Carazolol |
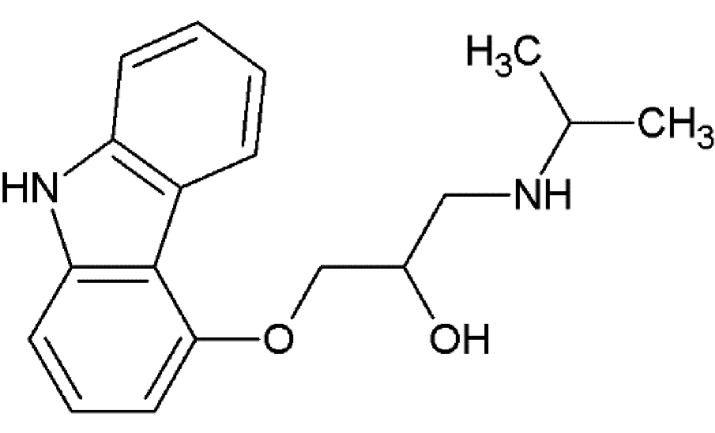
|
Inverse agonist | CHEMBL324665 | 298.4 | 0.114 | Sabio et al. (2008 ▸) |
| Timolol |
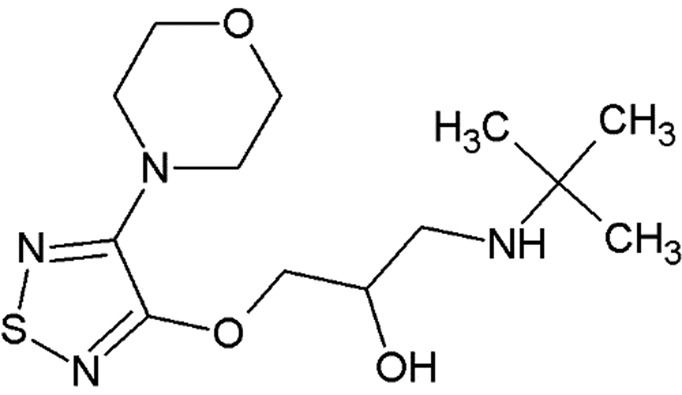
|
Inverse agonist | CHEMBL499 | 316.4 | 0.201† | |
| Propranolol |
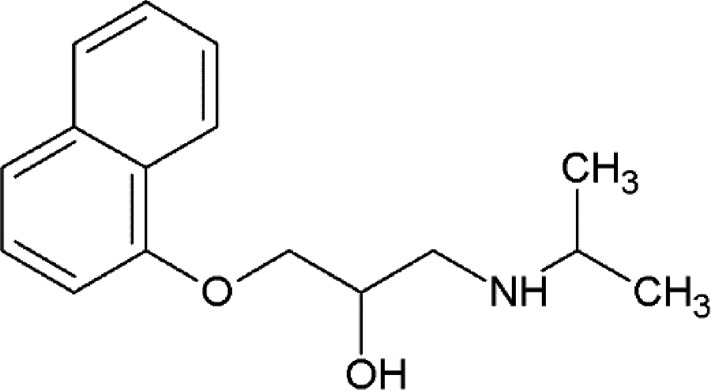
|
Inverse agonist | CHEMBL27 | 259.3 | 3.69 | Plazinska et al. (2014 ▸) |
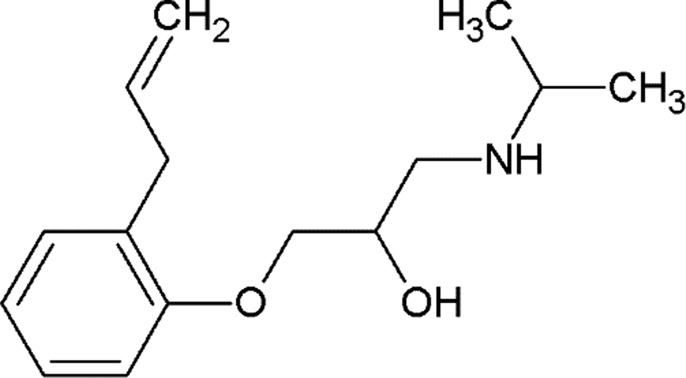
| Alprenolol |
|
Antagonist | CHEMBL266195 | 249.4 | 1 | Aristotelous et al. (2013 ▸) |
| Carvedilol |

|
β-Arrestin-biased agonist | CHEMBL723 | 406.5 | 0.166† | |
| Procaterol |

|
Agonist | CHEMBL160519 | 290.4 | 78 | Baker (2010 ▸) |
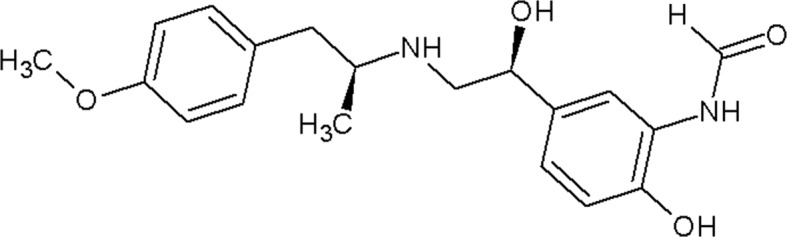
| Formoterol |
|
Agonist | CHEMBL3989798 | 344.4 | 23 | Baker (2010 ▸) |
Values are from the DrugMatrix Database (https://ntp.niehs.nih.gov/drugmatrix/index.html).
