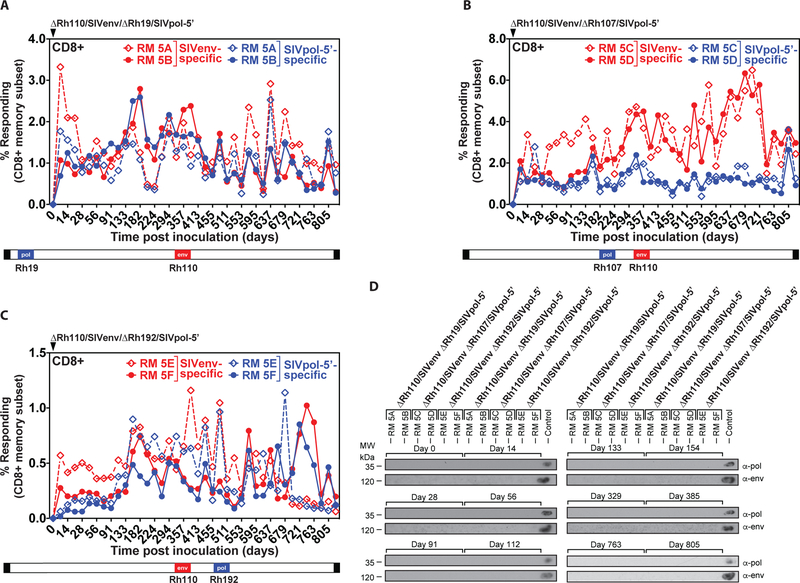
Figure 5: Genetic stability of Rh110 (pp71)-deleted RhCMV vectors in the setting of superinfection.
Two naturally RhCMV-infected RMs were each inoculated with 107 PFU of each dual insert-expressing ΔRh110/SIVenv/ΔRh19/SIVpol-5’ (A), ΔRh110/SIVenv/ΔRh107/SIVpol-5’ (B), or ΔRh110/SIVenv ΔRh192/SIVpol-5’ (C) and flow cytometric ICS was used to follow the SIVenv- and SIVpol-specific CD8+ T cell responses in peripheral blood, as described in Fig. 2. Dashed and solid lines each delineate the individual RM among the RM pairs given each vector. The relative position of the SIV antigens replacing endogenous genes in the viral genome is shown schematically below the graphs. CD4+ T cell responses from the same RM are shown in fig. S9. (D) Urine samples from the indicated time points post-vector inoculation were analyzed for vector shedding by viral co-culture followed by western blot analysis of SIVpol-5’ (top panels) or SIVenv (bottom panels) expression. Urine from RMs that previously received 68–1/SIVpol-5’ and 68–1/SIVenv vectors was included as a positive control.