Figure 1.
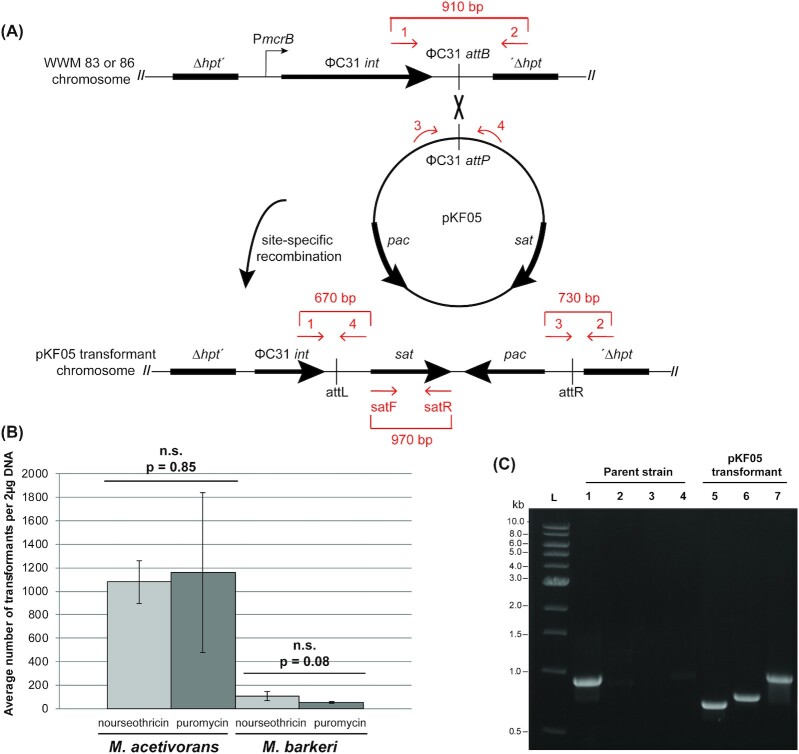
The streptothricin acetyltransferase (sat) gene is a useful positive selectable marker for Methanosarcina transformations. (A) A schematic of Streptomyces ΦC31 integrase-mediated site-specific recombination of pKF05 into M. acetivorans (WWM83) or M. barkeri (WWM86) chromosomes. ΦC31 integrase-mediated chromosomal integration of pKF05 introduces the selectable markers pac and sat for selection of Methanosarcina transformants. Primers used to verify pKF05 transformants are represented by the red arrows in the schematic. (B) Comparison of pKF05 transformation efficiency under puromycin or nourseothricin selection in M. acetivorans and M. barkeri. Triplicate transformation experiments with pKF05 were completed with M. acetivorans (WWM83) and M. barkeri (WWM86). Transformants were selected on both nourseothricin and puromycin plates. The number of colonies that arose was counted to determine the transformation efficiency: 1157 ± 681 and 1078 ± 182 transformants on puromycin and nourseothricin, respectively, for M. acetivorans and 55 ± 7 and 106 ± 37 on puromycin and nourseothricin, respectively, for M. barkeri. Error bars represent the standard deviation of three independent transformations and P-values were calculated using two-tailed unpaired t-tests (α = 0.01). Importantly, no colonies were observed for controls lacking DNA. (C) PCR verification of Methanosarcina pKF05 transformants. pKF05 transformants produced 670, 730, or 970 bp PCR products from reactions containing primers 1 and 4 (Lane 5), 2 and 3 (Lane 6) or satF and satR (Lane 7), respectively, while the parent strain only produced a 910 bp PCR product from reactions containing primers 1 and 2 (Lane 1) but no products with the other three primer pairs (Lanes 2–4).