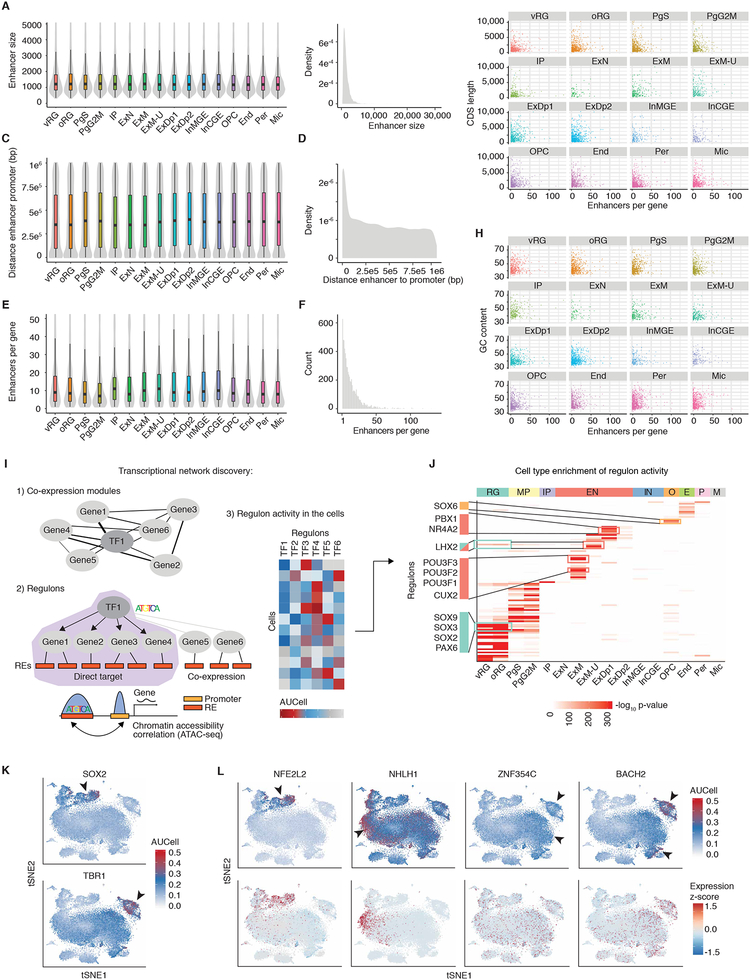
Figure 5. Transcriptional network discovery.
(A to H) Regulatory elements for cell-type specific genes. (A) Enhancer size by cell type. Enhancers are assigned to cell types by cell type enriched genes. (B) Density plot of enhancer sizes that are assigned to specific cell types. (C) Distance (base pairs) from enhancer to promoter by cell type. (D) Density plot of distance (base pairs) from enhancer end to promoter start that are assigned to specific cell types. (E) Enhancers per gene by cell type. (F) Histogram of enhancers per gene that are assigned to specific cell types. (G) Number of enhancers per gene versus CDS length of the gene by cell type. (H) Number of enhancers per gene versus GC content of the gene by cell type. (I) Schematic showing the computational approach used for transcriptional network discovery with the SCENIC pipeline (see materials and methods). 1) Co-expression modules between transcription factors and candidate genes are constructed. 2) Genes in co-expression modules are then pruned to genes which are inferred to be direct targets of the transcription factor, making a regulon. Direct targets are determined by the presence of the transcription factors binding motif in the regulatory elements associated with that gene. 3) The activity of each regulon is then assessed in each cell. (J) Cell type enrichment of regulon activity. Each regulon was scored as active or inactive for each cell, and cluster enrichment was then determined by Fisher’s test. Color indicates FDR-corrected -log10 p-value. (K) SCENIC regulon activity in each cell (AUCell) for the indicated TF plotted on tSNE. (L) TFs with previously uncharacterized cell type or cell subtype specific activity. Regulon activity in each cell (AUCell) for the indicated TF (top panels) or expression of the TF plotted on tSNE (bottom panels).