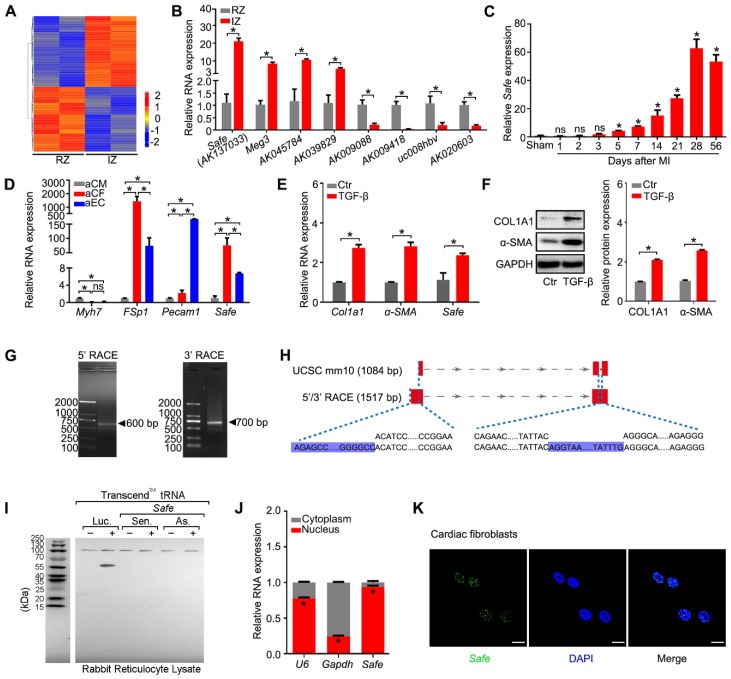
Figure 1.
Safe is a fibroblast-enriched lncRNA and associated with cardiac fibrosis. (A) Heatmap showing 389 lncRNAs differentially expressed in tissues from the infarction zone (IZ) and non-infarcted remote zone (RZ) at day 14 post MI (fold ≥ 2, p<0.05). (B) qRT-PCR validation of four upregulated lncRNAs and four downregulated lncRNAs in cardiac samples from IZ and RZ of infarcted hearts (n=3). (C) qRT-PCR analysis of Safe expression in healthy (sham) or infarcted myocardium at indicated days post MI (n=3). (D) qRT-PCR detection of Safe expression in adult cardiomyocytes (aCM), adult cardiac fibroblasts (aCF) and adult endothelial cells (aEC). Myh7 (myosin heavy chain 7), FSp1 (fibroblast specific protein-1) and Pecam1 (Platelet endothelial cell adhesion molecule-1) were used as markers of aCM, aCF and aEC, respectively (n=3). (E) qRT-PCR detection of Safe, Col1a1 and α-SMA expression in TGF-β-treated cardiac fibroblasts (n=3). (F) Representative western blot analysis and relative densitometric quantification of COL1A1 and α-SMA protein levels in cardiac fibroblasts with TGF-β treatment (n=3). (G) Agarose gel electrophoresis of the 5' and 3' RACE amplification products. (H) Schematic presentation of full-length Safe showing the extended regions identified by RACE (blue). The full-length of Safe was 1517 base pairs with two exon regions, which was different from the sequence information provided by UCSC mm10 showing 1048 base pairs in length and three exon regions. (I) In vitro translation of Safe sense or antisense transcript. Luciferase (Luc) was used as a positive control. (J) Subcellular localization of Safe in cytoplasm and nucleus of cardiac fibroblasts (n=3). The gene U6 and Gapdh were respectively used as nuclear and cytoplasmic RNA markers. (K) Representative images of RNA FISH showing nuclear localization of Safe (green) in cardiac fibroblasts. The nucleuses were counterstained with DAPI (blue). Scale bar indicates 20 μm. Data are presented as mean ± SEM and analyzed using Student's t-test or one-way ANOVA; *p < 0.05, and ns, not significant.