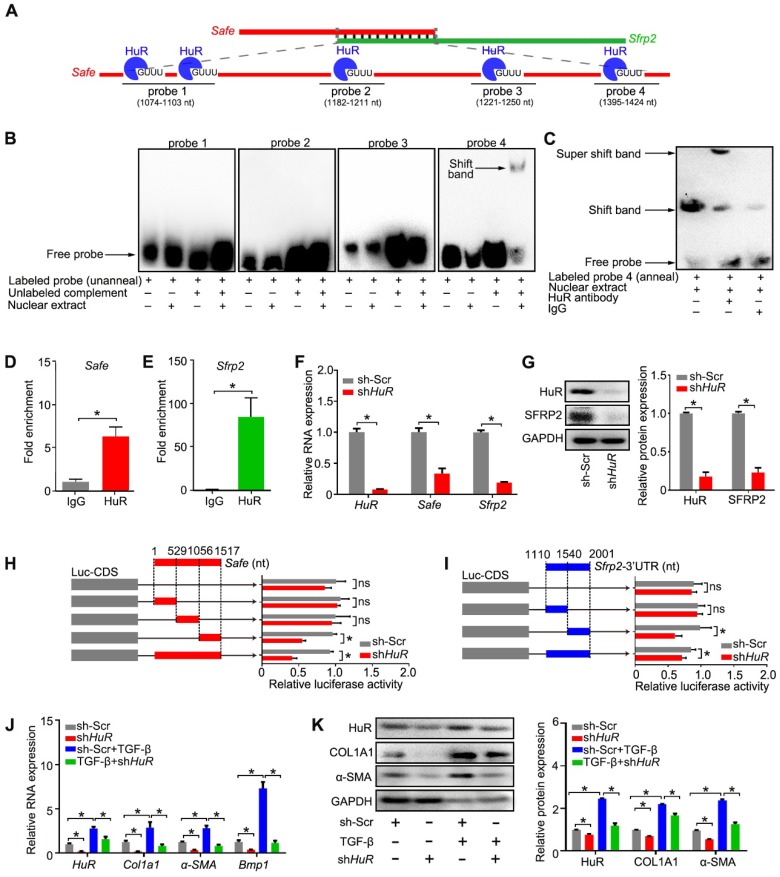
Figure 7.
Binding of HuR to the Safe-Sfrp2 duplex accelerates RNA stabilization of both Safe and Sfrp2. (A) Schematic presentation of predicated HuR binding sites and corresponding RNA probes for EMSA assay in the complementary region of Safe and Sfrp2 RNA. (B) EMSA assay indicating specific binding of nuclear proteins of cardiac fibroblasts to the 26-nucleotide RNA duplexes corresponding to nucleotide at the 1408 nt protein binding site of Safe. (C) EMSA supershift assay revealing the in vitro interaction between HuR protein and the 26-nucleotide RNA duplexes in nuclear extracts. RNA immunoprecipitation (RIP) assay using HuR antibody and IgG (isotype control) showing enrichment of Safe (D) and Sfrp2 (E) RNAs to HuR protein. (F) qRT-PCR analysis showing decreased expression of HuR, Safe and Sfrp2 in cardiac fibroblasts after HuR knockdown (n=3). (G) Representative western blot analysis and relative densitometric quantification of HuR and SRRP2 protein levels in cardiac fibroblasts with or without HuR inhibition (n=3). (H) Dual luciferase assay showing shHuR inhibited Firefly luciferase activities of pGL3-control vectors carrying the 3'-end of Safe (462-nucleotide in length) (n=3). (I) Dual luciferase assay showing shHuR inhibited Firefly luciferase activities of pGL3-control vectors carrying Sfrp2 3'-UTR (n=3). (J) qRT-PCR detection of HuR, Col1a1, α-SMA and Bmp1 in TGF-β-untreated and TGF-β-treated cardiac fibroblasts after HuR knockdown (n=3). (K) Representative western blot analysis and relative densitometric quantification of HuR, COL1A1 and α-SMA protein levels in sh-Scr or HuR-silenced cardiac fibroblasts after TGF-β treatment (n=3). All data are presented as mean ± SEM; Student's t-test or one-way ANOVA, *p < 0.05, and ns, not significant.