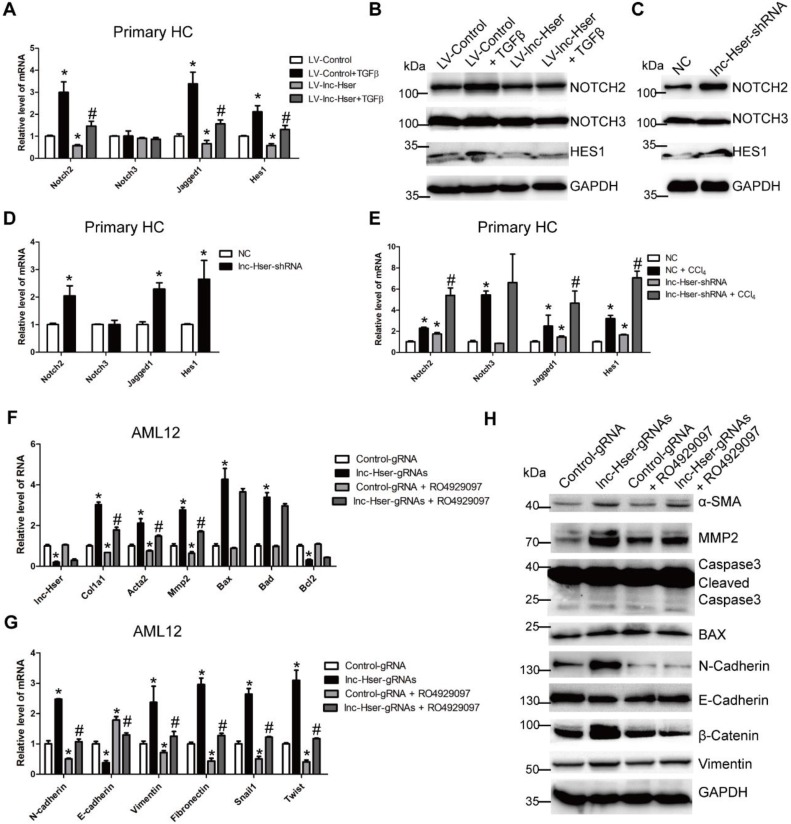
Figure 6.
lnc-Hser inhibits HCs EMT via the Notch pathway. (A, B) Primary HCs were infected with LV-lnc-Hser for 72 h and further treated with 10 ng/ml TGFβ for additional 24 h. The mRNA level of Notch2, Notch3, Jagged1 and Hes1 was detected by qRT-PCR (A). The protein level of NOTCH2, NOTCH3 and HES1 was detected by western blot. GAPDH was used as an internal control (B). (C) The protein level of NOTCH2, NOTCH3 and HES1 in primary HCs infected with lenti-lnc-Hser-shRNA or lenti-NC was detected by western blot. GAPDH was used as an internal control. (D) The mRNA level of Notch2, Notch3, Jagged1 and Hes1 in primary HCs infected with lenti-lnc-Hser-shRNA or lenti-NC was detected by qRT-PCR. (E) Mice were treated with oil in combination with injection of lenti-NC (Negative Control, n = 10), or CCl4 in combination with injection of lenti-NC (NC + CCl4, n = 10), or oil in combination with injection of lenti-lnc-Hser-shRNA (lnc-Hser-shRNA, n = 10), or CCl4 in combination with injection of lenti-lnc-Hser-shRNA (lnc-Hser-shRNA + CCl4, n = 10). qRT-PCR analysis of Notch2, Notch3, Jagged1 and Hes1 in the primary HCs isolated from mice in each group. (F-H) lnc-Hser was stably knocked down by the CRISPR/Cas9 system with guide RNA pairs in AML12 cells. The γ-secretase inhibitor RO4929097 was used to treat lnc-Hser-silenced AML12 cells for 24 h. The expression of lnc-Hser, pro-fibrogenic, apoptosis-related and EMT-related genes was detected by qRT-PCR (F, G) and western blot. GAPDH was used as an internal control (H). The data are expressed as the mean ± SD for at least triplicate experiments. *p<0.05 for stands vs LV-Control, NC or Control gRNA. #p<0.05 stands for vs LV-Control + TGFβ, NC + CCl4 or lnc-Hser-gRNAs.