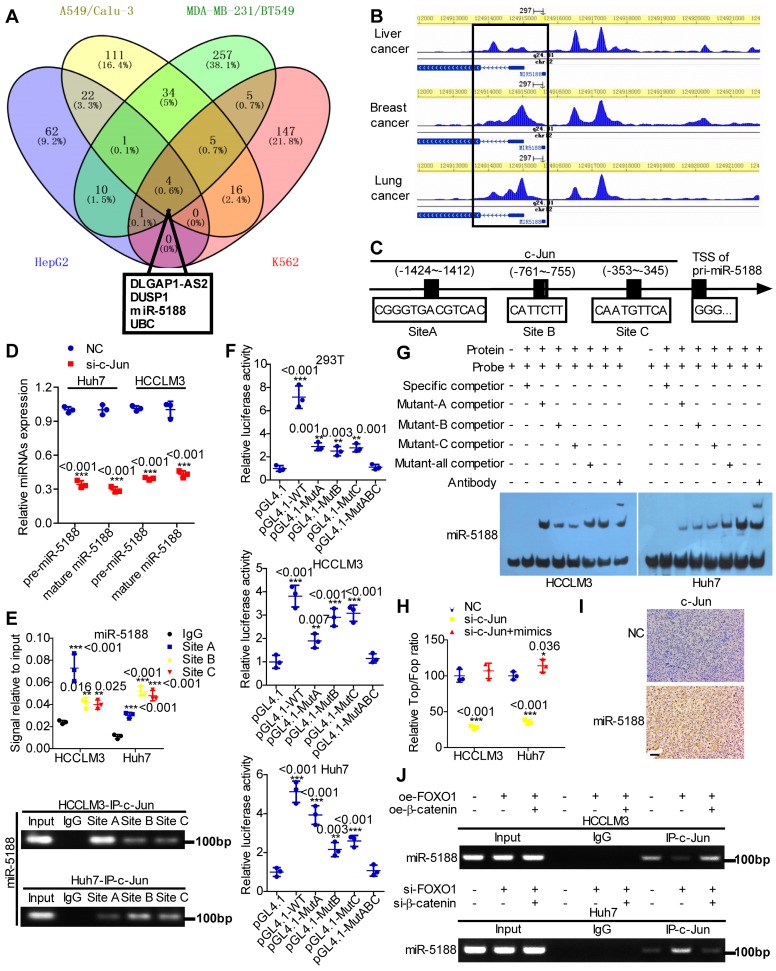
Figure 5.
c-Jun transcriptionally promotes miR-5188 expression to form a positive regulatory loop. (A) Venn diagram analysis was used to screen c-Jun-regulated downstream effectors among the top putative targets based on the Cistrome Data Browser. (B) ChIP-seq binding peaks were searched by the Cistrome Data Browser. (C) Bioinformatics analysis was utilized to predict c-Jun-binding sites within the promoter of miR-5188. (D) QPCR was used to examine pre-miR-5188 and mature miR-5188 levels in c-Jun-silenced HCC cells and control cells (n=3 independent experiments, Student's t-test). (E) Chromatin immunoprecipitation (comparison of all groups vs. the IgG group) (n=3 independent experiments, one-way ANOVA) was used to identify c-Jun binding to the miR-5188 promoter. (F) Luciferase reporter assays (comparison of all groups vs. the control group) (n=3 independent experiments, one-way ANOVA) were performed to confirm c-Jun binding to the miR-5188 promoter. (G) Protein-DNA interactions between c-Jun and the miR-5188 promoter were determined using electrophoretic mobility shift assays. (H) TOP/FOP luciferase reporter assays were performed to detect Wnt/β-catenin signaling activity (n=3 independent experiments, one-way ANOVA). (I) Immunohistochemical analysis was used to detect c-Jun expression in xenograft tumors derived from Huh7 cells after stable miR-5188 overexpression and control cells (scale bar: 20 μm) (n=5). (J) Chromatin immunoprecipitation analysis assessed c-Jun binding to the miR-5188 promoter in FOXO1-overexpressing HCCLM3 cells, FOXO1-overexpressing HCCLM3 cells with β-catenin overexpression, FOXO1-silenced Huh7 cells, FOXO1-silenced Huh7 cells with β-catenin knockdown, and the corresponding control cells. All data are presented as the mean ± SD. Experiments were repeated three times.