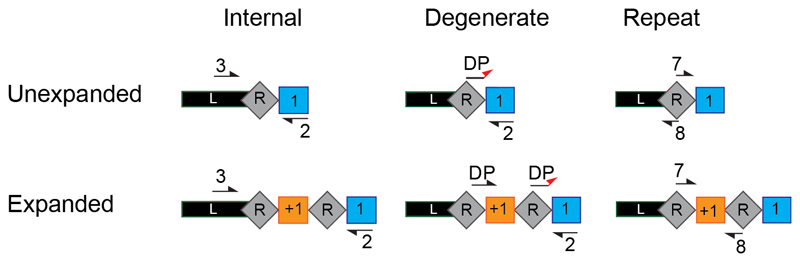
Figure 5. Primer design options for re-amplification of expanded CRISPR arrays.
Schematic of the three possible primer options for the second PCR amplification step. The internal primers (left) bind in spacer 1 (primer #2) and the leader sequence (#3) internally of primer #1 used in the first PCR step before size selection. The degenerate primers (middle) consist of 3 forward primers (DP) that anneal their 3’ nucleotide only with new spacers (orange) starting with a nucleotide different from the original spacer (blue), in combination with #2. The repeat primers (right) bind within the repeat (#7 and #8) orientated such that a product is only amplified when two repeats are present i.e. the array is expanded.