Fig. 1. SWEET11–SWEET13–SWEET14 EBE pentagon and PCR detection of SWEET induction.
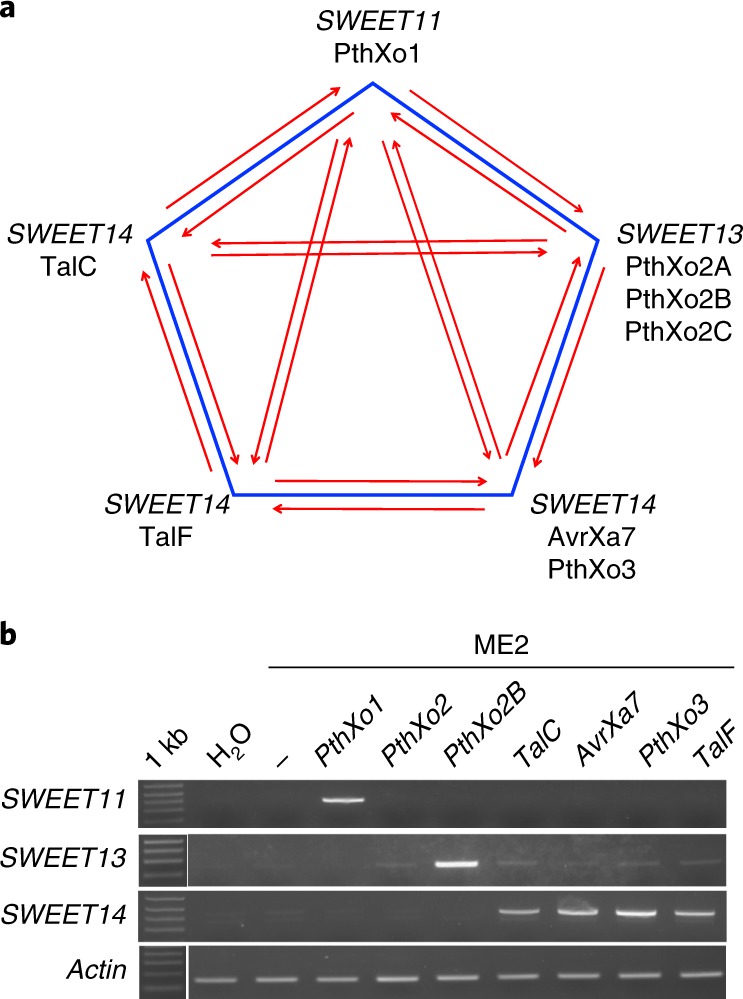
a, Arrows indicate which TAL effectors can overcome a particular resistance mechanism by activating any of the other SWEET genes or by activating the same SWEET gene via targeting another EBE in the same promoter. For example, xa13-based resistance (resulting from a variant in the SWEET11 promoter that is not recognized by PthXo1) can be overcome by other TAL effectors (e.g., PthXo2 and PthXo3) that target the EBEs in another SWEET gene promoter or, in the case of SWEET14, target different EBEs in the same promoter. b, Example of SWEET gene induction as detected by RT–PCR using the SWEETup primer set. RT–PCR products are shown for the SWEET11, SWEET13 and SWEET14 genes in Kitaake infected by Xoo strain ME2 lacking a SWEET-targeting TAL effector and ME2 transformed with plasmids encoding PthXo1 (targeting SWEET11), PthXo2 or PthXo2B (both targeting SWEET13) and PthXo3, TalC, TalF or AvrXa7 (all targeting SWEET14). Actin served as a control. Leaves were infected using leaf-clipping assays; scissors dipped in water served as an additional negative control. The experiment was repeated twice independently with similar results.
