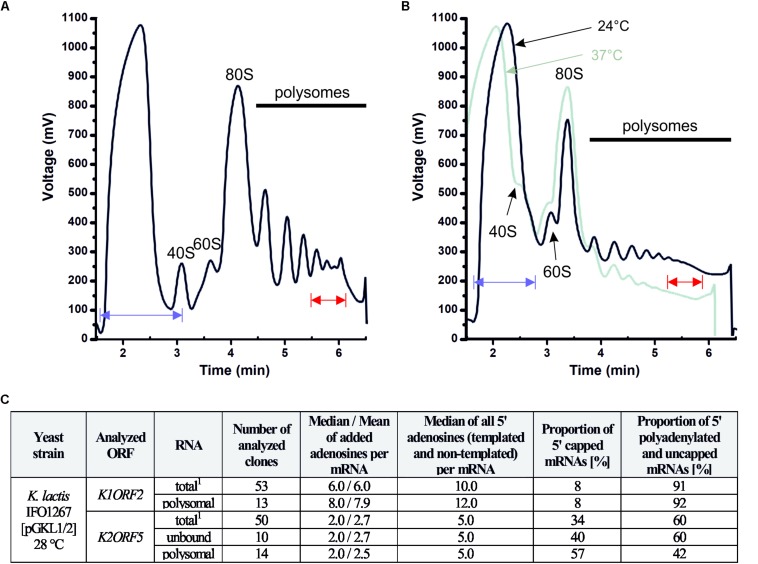
FIGURE 11.
The 5′ polyadenylated and uncapped pGKL mRNAs are loaded onto yeast high polysomes. (A) Analysis of polysomes from K. lactis IFO1267 grown exponentially in YPD medium at 28°C. The fraction of high polysomes (depicted in red) and the non-polysomal fraction (unbound, depicted in blue) were used for the mRNA purification and 5′ RACE analysis of pGKL mRNAs. (B) Analysis of polysomes from the S. cerevisiae CWO4 cdc33-42ρ0 [pGKL1/2] strain grown in YPD medium in parallel at permissive (24°C, black curve) and restrictive (37°C, green curve) temperatures. Overall cellular translation was substantially decreased in the absence of functional eIF4E (green curve). The fractions of high polysomes (depicted in red) and non-polysomal fractions (depicted in blue) from cultures grown at the permissive (24°C) and non-permissive (37°C) temperatures were used for the total RNA purification and 5′ RACE analysis of pGKL mRNAs (Figures 12, 13). (C) Tabular summary of the 5′ RACE results from mRNAs purified from unbound and high polysome fractions in the experiment “A”. Messenger RNAs of each ORF subjected to 5′ RACE analysis are characterized by the following data: (1) ORF name; (2) source of purified mRNA; (3) total number of analyzed clones; (4) median and mean of 5′ added adenosines per mRNA molecule; (5) median of all 5′ adenosines (templated and non-templated) per mRNA molecule; (6) proportion of 5′ capped mRNAs [in %]; (7) proportion of 5′ polyadenylated and uncapped mRNAs [in %]. 40S, eukaryotic small ribosomal subunit; 60S, eukaryotic large ribosomal subunit; 80S, eukaryotic ribosome (monosome).