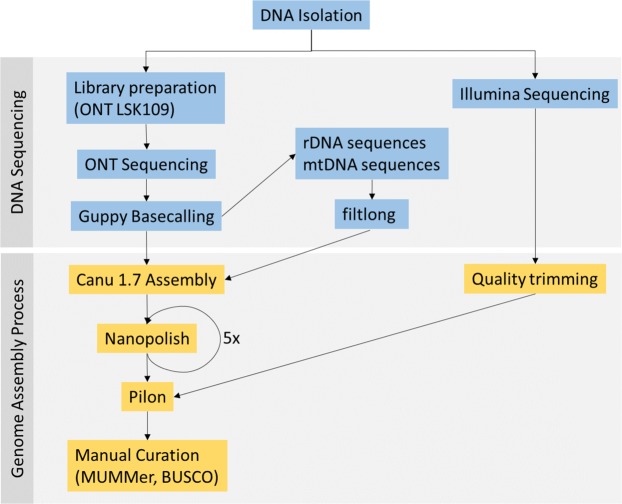
Figure 6.
Genome Assembly Workflow. Total DNA was isolated followed by library preparation using the ONT LSK109 kit and sequencing. The other part of the DNA sample was used for Illumina sequencing. Reads of the ONT sequencing were base called using Guppy. Before assembly of the nuclear genome, reads of the mitochondrial genome and the extrachromosomal rDNA plasmid were removed by aligning them to N. gruberi reference sequence. The nuclear genome was then assembled using Canu 1.7 followed by 5 rounds of Nanopolish v0.11.0 and one round of Pilon v1.22 in combination with trimmed high-quality Illumina data. The polished assembly was then manually curated to remove redundant sequences based on MUMmer v4.0.0 alignments and duplicated BUSCOs. The mitochondrial genome and the rDNA plasmid were assembled separately. Therefore, reads were quality filtered using filtlong followed by assembly using Canu and polishing using Nanopolish and Pilon as described above.