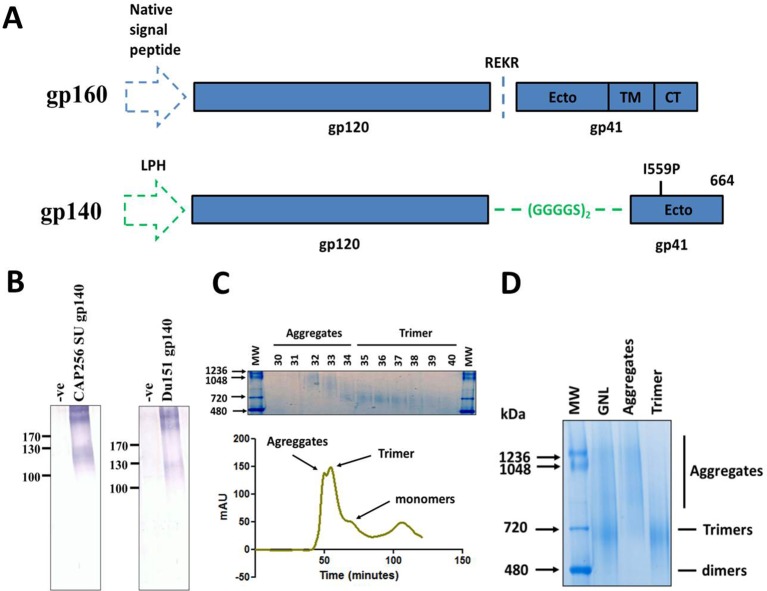
Figure 1.
Design, expression and purification of HIV Env gp140 antigens. (A) Schematic of the coding sequence for the native HIV-1 gp160 gene (top) and the soluble gp140 antigen (bottom). The gp120 and gp41 portions of the proteins are delineated with either the native cleavage sequence (REKR) or flexible linker peptide (GGGGS)2 at the interface of the two subunits. The location of the I559P helix breaking mutation and amino acid residue 664, where the coding sequence was terminated, is reflected for the gp140 antigen. The native and LPH (leader peptide heavy chain) signal sequences are shown by the blue and green dashed arrows respectively. (Ecto = ectodomain, TM = transmembrane, CT = cytoplasmic tail). (B) Western blot confirming expression of CAP256 SU gp140 (left) and Du151 gp140 (right) in crude leaf homogenate, 5 days post infiltration. Blots were probed with polyclonal goat anti-HIV-1 gp120 antibody. The negative controls comprise of leaf tissue agroinfiltrated with recombinant A. tumefaciens AGL1 that has been transformed with the empty pEAQ-HT expression vector (-ve). Both CAP256 SU Env and Du151 Env contain 29 putative N-glycosylation sites. (C) Superdex 200 Hiload 16/600 elution profile of fractionated CAP256 SU Env species following affinity chromatography. The identities of the different protein species are indicated on the profile. A Coomassie-stained BN-PAGE gel of the fractionated material is shown above the graph indicating the fractions that comprise the aggregates and presumed trimer peaks. (D) Coomassie stained BN-PAGE gel containing the pooled and concentrated CAP256 SU Env samples corresponding to peaks in the elution profile peaks. An aliquot of the affinity-purified protein (GNL), prior to SEC, was resolved alongside aggregate and trimer samples for comparison. The trimers indicated in both panels (C) and (D) are based on the SEC elution profiles and BN-PAGE migration of other HIV-1 gp140 antigens in the published literature.