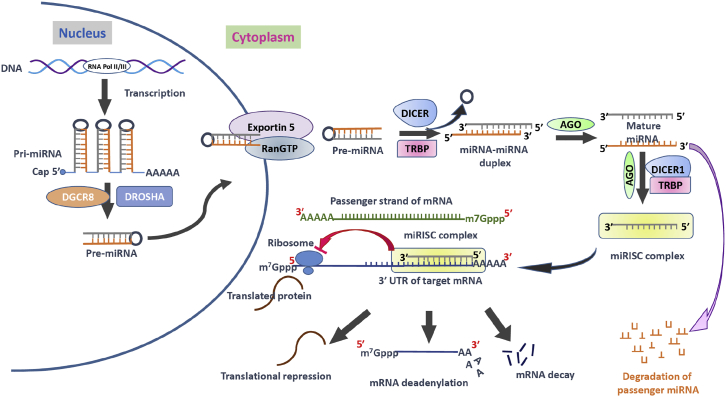
Figure 1.
Mechanism of MicroRNA Processing and Their Inhibitory Mechanism
The microRNA (miRNA) processing pathway begins with transcription of their genes with the help of RNA polymerase II (Pol II) or polymerase III (Pol III) to produce pri-miRNAs in the nucleus. Then a microprocessor complex, composed of RNA-binding protein DGCR8 and type III RNase Drosha, cleaves pri-miRNA into a ∼85-nt stem-loop structure called pre-miRNA. The exportin 5-RAN/GTP complex mediates the transport of pre-miRNA from the nucleus into the cytoplasm. The RNase DICER in complex with double-stranded RNA-binding protein TRBP cleaves the pre-miRNA hairpin to a ∼20- to 22-nt miRNA/miRNA duplex. After the duplex is unwound, the functional strand of the mature miRNA (the guide strand) is loaded into the miRISC-containing DICER1, TRBP, and Argonaute (AGO) proteins. This miRISC silences/inhibits the target mRNAs expression/function through mRNA cleavage, translational repression, or deadenylation. The passenger strand of the miRNA is degraded. AGO, Argonaute proteins; DGCR8, DiGeorge syndrome critical region gene 8; m7G cap, 7-methylguanosine; miRISC, miRNA-induced silencing complex; miRNA, microRNA; pre-miRNA, miRNA precursor; pri-miRNA, primary miRNA; RAN-GTP, Ras-related nuclear protein coupled with guanosine-5′-triphosphate; TRBP, transactivating response RNA-binding protein.