Figure 2.
Benchmarking and Remodeling of mtPPIs in ECSCs and DNLCs
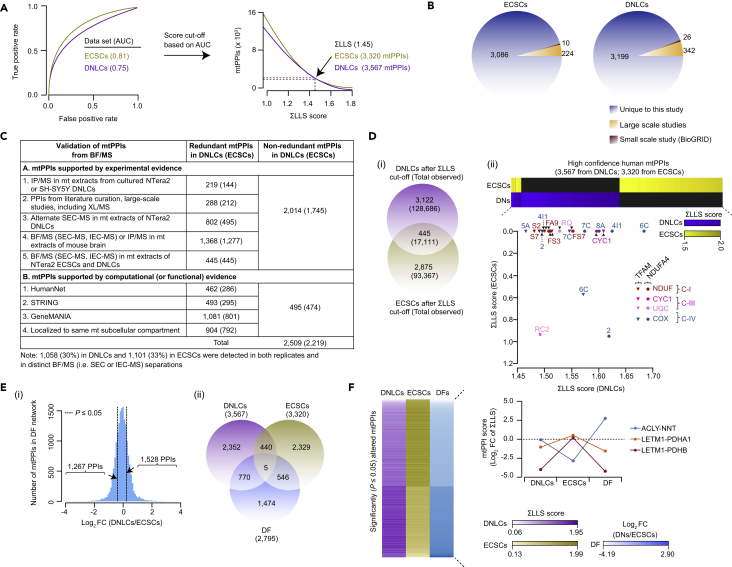
(A) Performance measures using AUC (area under the receiver operating characteristic curve) analysis based on estimates of true- and false-positive rates for scored mtPPIs against reference CORUM protein complexes containing mtPPIs (top). Cumulative ΣLLS score (bottom; threshold based on AUC) was computed by combining LLS score derived from SEC and IEC fractions.
(B) Overlap of high-confidence mtPPIs from ECSC and DNLC networks against the large- or small-scale (curated in BioGRID database) studies.
(C) Experimental and computational evidence supporting mtPPIs from each cell state.
(D) High-confidence (and total) mtPPIs common or specific to ECSC or DNLC networks (i). Association of CI subunit (NDUFA4) or TFAM with the components of the respirasome (ii) along with their ΣLLS scores in two static networks.
(E) Z score distribution (i) of log2 fold change (FC) for differential (DF) PPIs filtered at p value ≤ 0.05 (|Z score| ≥ 1.96) with tails indicating significant interactions. Venn diagram (ii) showing the overlap of significant mtPPIs in static and DF networks.
(F) Heatmap displaying significantly altered mtPPIs during differentiation (left) is shown with illustrative examples (right) for static and DF profiles.
See also Figures S2 and S3, and Table S2.