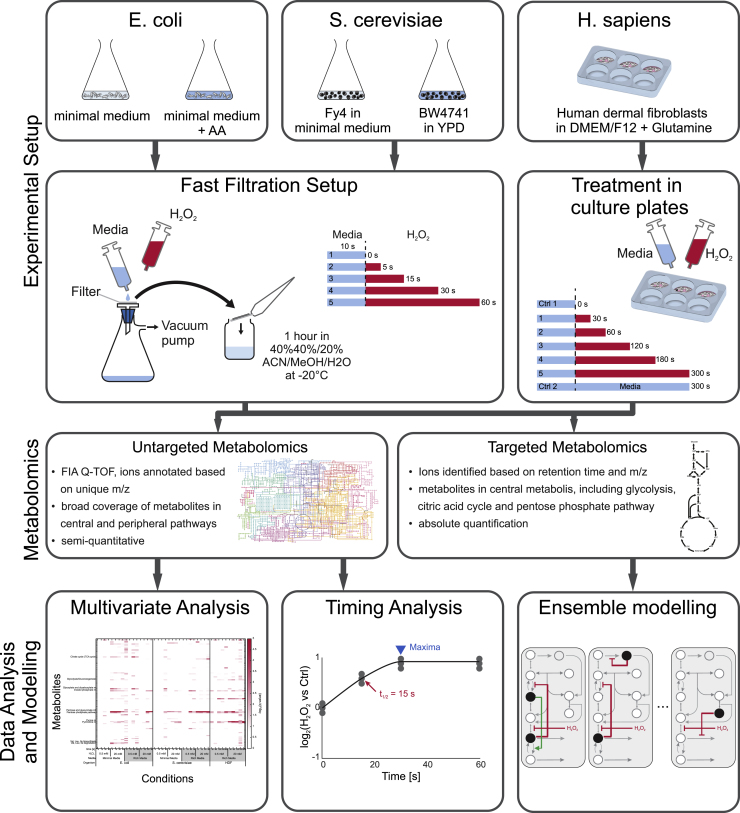
Figure 1.
Experimental— – Computational Workflow
Triplicate cultures of E. coli and S. cerevisiae were grown in rich and minimal medium and human dermal fibroblasts in rich medium. Mid exponential growth phase cultures of microbes were transferred to a filter and for 10 s perfused with the cultivation medium and then with the same medium but with either a low (0.5 mM) or high (20 mM) dose of H2O2. The mammalian stress experiments were performed in liquid culture through addition of H2O2 dosage. Culture aliquots were immediately transferred into approximately −20°C cold quenching/extraction liquid and prepared for mass spectrometric analysis of the intracellular metabolome. Using the data from the untargeted metabolomics measurements, we performed multivariate analysis and timing analysis. To systematically map all metabolite-enzyme interactions and their functional relevance, we developed kinetic models of glycolysis and the PP pathway for all species and conditions. Ensembles of models with different putative regulatory interactions were then tested for their ability to capture the dynamics of eight metabolites in central metabolism.