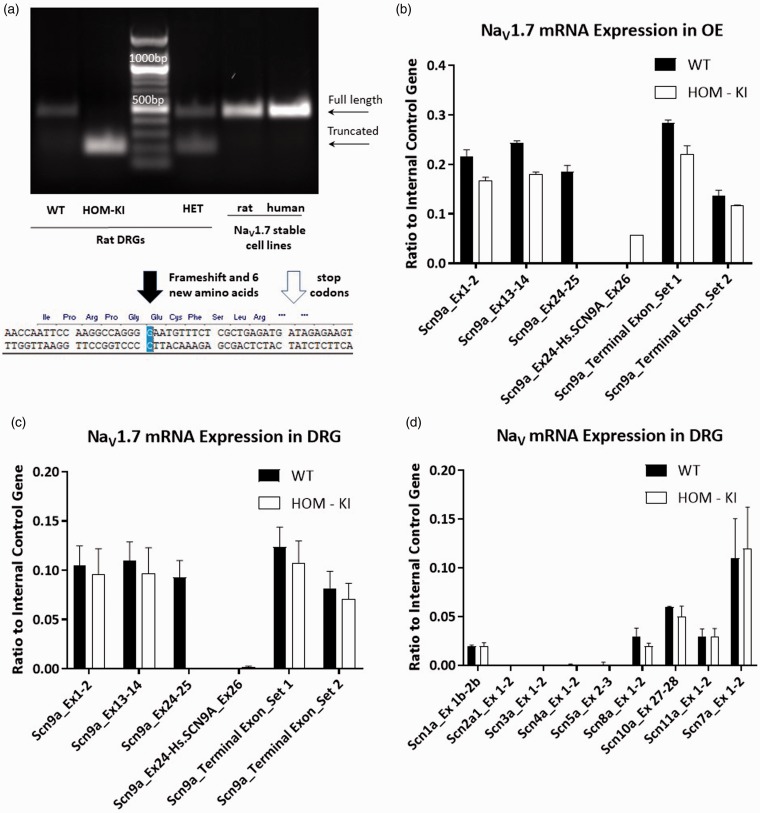
Figure 6.
NaV1.7 transcript analysis in sensory tissues. (a) RT-PCR analysis of adult WT vs. HOM-KI DRGs and reference human and rat NaV1.7 stable HEK293 cell lines. Full-length bands were detected in WT, HET tissue samples as well as in positive controls (stable cell lines) (463 bp) in addition to an unexpected truncated band in HOM-KI and HET samples only (193 bp). Sequence of truncated RT-PCR band revealed splicing out of human exon 26 plus an additional guanine nucleotide from rat exon 26 with a resulting frameshift (black arrow), insertion of six novel amino acids, and pre-mature stop codons (white arrow). (b and c) NaV1.7 amplicons in WT and HOM-KI using primers positioned in various rat or human NaV1.7 exons upstream and downstream of the insertion in olfactory epithelium (OE) (b) and DRG (c). RT-PCR products were detected for rat NaV1.7 amplicons prior to and following human exon 26 in WT and HOM-KI tissue. An amplicon between rat exon 24 and human exon 26 was nominally detected in DRG tissue but was present in OE tissue in HOM-KI rats. (d) RT-PCR of NaV isoforms in DRG tissue. No compensatory expression changes were observed for non-NaV1.7 isoforms in HOM-KI rats. Data are mean ± SEM, four rats of each genotype (two males and two females) were evaluated. HOM-KI: rats homozygous for the knock-in allele; WT: wild type; OE: olfactory epithelium; DRG: dorsal root ganglia.