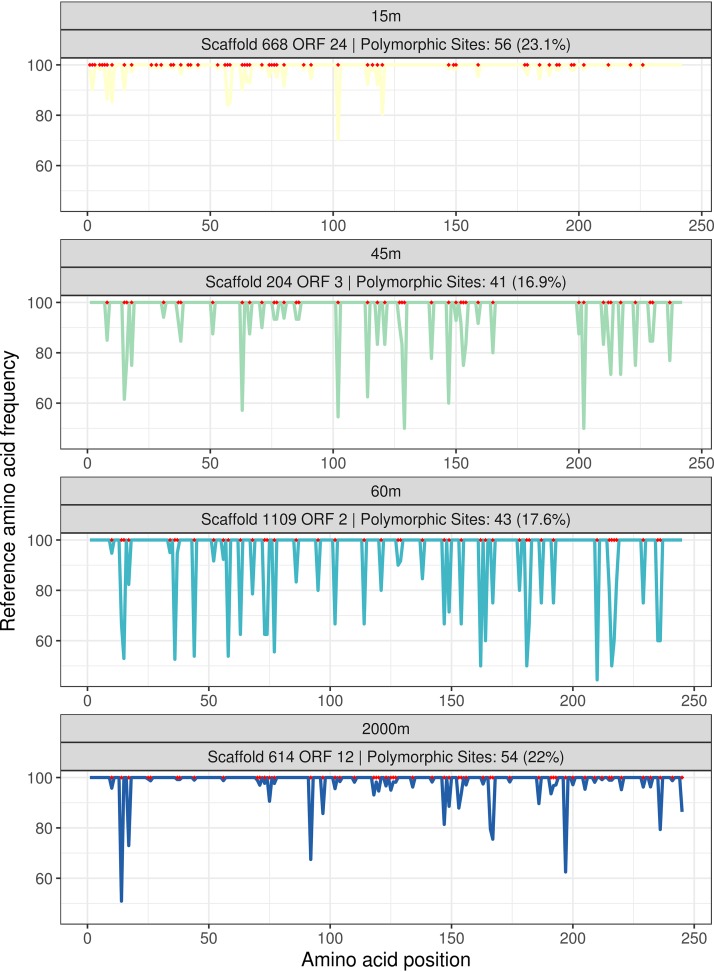
FIG 5.
Occurrence of nonsynonymous mutations among a group of homologous tail proteins. These proteins are homologous and annotated as tail proteins (which are involved in host recognition). Each of them (ORF 24 from scaffold 668 in the 15-m virome, ORF 3 from scaffold 204 in the 45-m virome, ORF 2 from scaffold 1109 in the 60-m virome, and ORF 12 from scaffold 614 in the 2,000-m virome) was retrieved from scaffolds assembled at different depths, thus allowing for the comparison of the patterns of accumulation of nonsynonymous mutations at each analyzed depth among homologous proteins. In the line plots of the bottom panel, the x axis depicts the amino acid position along proteins, and the y axis depicts the frequency of the reference amino acid (i.e., the most frequent) among the reads from each sample. The red dots along the x axis indicate which sites throughout the protein are polymorphic sites. Valleys in the plot represent areas that concentrate nonsynonymous mutations, possibly driven by positive selection favoring mutations that modify or expand host range.