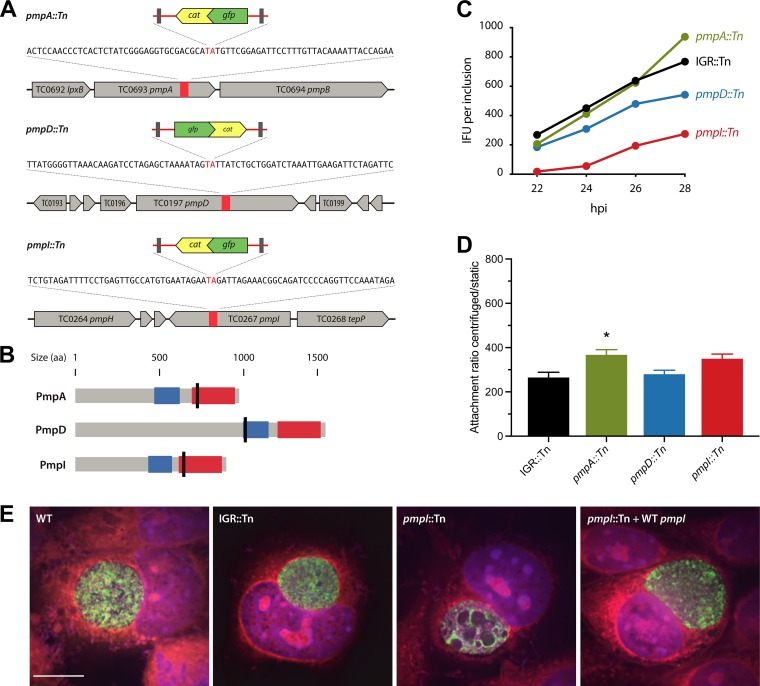
FIG 5.
In vitro phenotypes of C. muridarum pmp mutants. (A) Chromosomal locations of transposon insertions in pmp genes. The orientations of inserts are indicated by the gene directionality. (B) Predicted protein topologies of Pmp proteins. Transposon insertion sites are marked with black lines; blue, Pmp middle domain; red, autotransporter domain. (C) Inclusion burst sizes of pmpI::Tn, pmpA::Tn, and pmpD::Tn strains versus an IGR::Tn control strain. McCoy cells infected with the indicated strains were harvested at the times postinfection indicated, and the amount of infectious progeny in each sample was determined by infecting fresh McCoy cells, followed by IFU assays. (D) Attachment of the indicated strains to McCoy cells was determined by inoculating the cells under centrifugation-aided and static conditions. The ratios of centrifugation to static conditions were computed. The data represent means, and the error bars indicate standard deviations. Statistical comparisons between transposon mutants and the IGR::Tn control were analyzed using one-way ANOVA with Dunnett’s test for multiple comparisons. *, P < 0.05. (E) Confocal immunofluorescence analysis of inclusions containing WT C. muridarum, the IGR::Tn or pmpI::Tn mutant, and the complemented pmpI::Tn-plus-WT pmpI strain analyzed at 24 hpi. Green, anti-LPS; blue, DAPI; red, Evans blue. Scale bar, 10 μm.