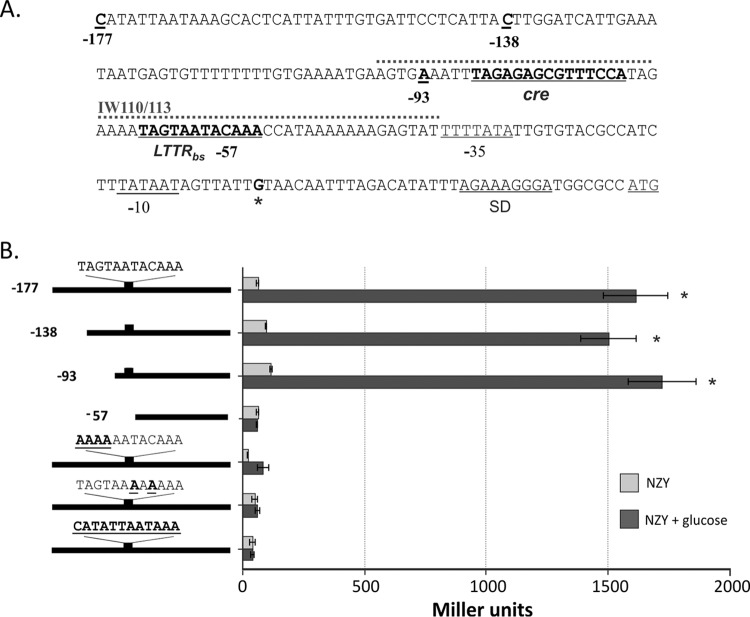
FIG 1.
Identification of cis-acting elements in the cidA promoter region. (A) Nucleotide sequence of the cidA promoter. The −35 and −10 elements, Shine-Dalgarno (SD) sequence and translational start codon are underlined. The transcriptional start point is marked by an asterisk and presented in bold. The positions of truncated DNA fragments used for the deletion analysis of cidA promoter activity, the putative CidR binding site, and identified catabolite responsive elements (cre) are underlined and presented in bold. A 60-bp biotin-labeled DNA fragment used in EMSAs (IW110/113) is marked by a dotted line. (B) Deletion and mutational analysis of cidA promoter activity. Wild-type S. aureus cells (strain UAMS-1) containing lacZ reporter fusions with truncated cidA promoter fragments or the full-length cidA promoter with mutations in the putative CidR-binding site were grown under inducing conditions for 6 h and subsequently assayed for β-galactosidase activity. The length of the cidA promoter fragments used for the deletion analysis and the wild-type sequence of the CidR-binding site with generated mutations are shown in bold and underlined on the left. The results are representative of at least three independent experiments. Statistical significance in β-galactosidase activity between the wild‐type strain containing cidA promoter fragments in the presence or absence of glucose (*) was determined by Student's t test (P ≤ 0.01).