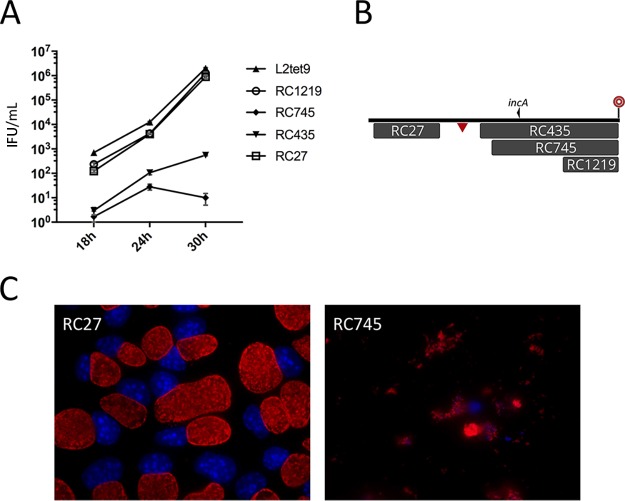
FIG 4.
Examination of recombinant clones that express a growth defect. (A) Inclusion-forming units were determined at three time points following inoculation of McCoy cells with a set of four recombinants plus the C. trachomatis L2tet9 parent. The numbers on the vertical axis are inclusion-forming units per milliliter of lysed, infected cells. Monolayers were inoculated with approximately 30,000 IFU per ml. The error bars represent standard deviations. (B) Map of the recombinants tested in the growth experiments. This is a subset of the library of recombinants shown in Fig. 2 and includes recombinants from regions D (RC27) and E (RC435, RC745, and RC1219) in Fig. 2. The locations of incA and the 32-bp recombination hot spot can be identified here and in Fig. 2 for positioning. The triangle indicates the homologous position in C. muridarum that is targeted in transposon mutant CM013, for which no syntenic recombinants were generated. For scale, the C. muridarum insert shown for recombinant RC27 is 52 kb. (C) Immunofluorescence microscopic images of a methanol-fixed McCoy cell recombinant progeny strain with a wild-type inclusion morphology phenotype (RC27) and a recombinant expressing the early-lysis phenotype (RC745). The cells were fixed at 30 h and had identical multiplicities of infection. Red, MOMP; blue, DNA.