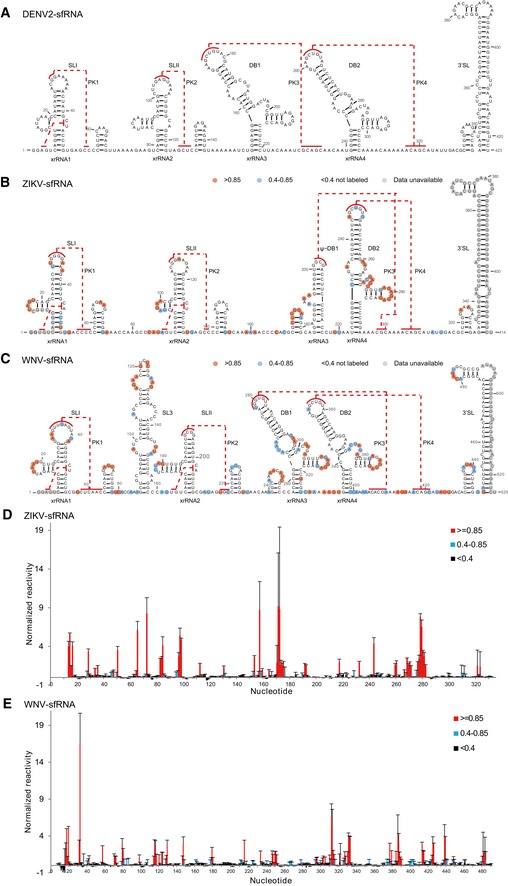
Figure 1. Secondary structures of the complete sfRNAs.
-
A–CSecondary structure of the complete sfRNAs of DENV2 (A), ZIKV (B), and WNV (C). The stem‐loop structures (SLI and SLII), SL3 of WNV, pseudo‐dumbbell (ψ‐DB1) structure of ZIKV, duplicated dumbbell structures (DB1, DB2), and the essential 3′ SL are indicated. Sequences involved in pseudoknots (PK1, PK2, PK3, PK4) formation are indicated with red lines.
-
D, ESHAPE analysis of the complete sfRNAs of ZIKV (D) and WNV (E). Nucleotides with SHAPE reactivity greater than 0.85 are labeled in red, and moderately reactive site (0.4–0.85) is labeled in cyan. Unlabeled sites have low or no SHAPE reactivity (< 0.4). The SHAPE reactivity was also annotated onto the corresponding secondary structures in (B) and (C). Nucleotides which SHAPE reactivity was not determined are labeled in gray in (B) and (C).
