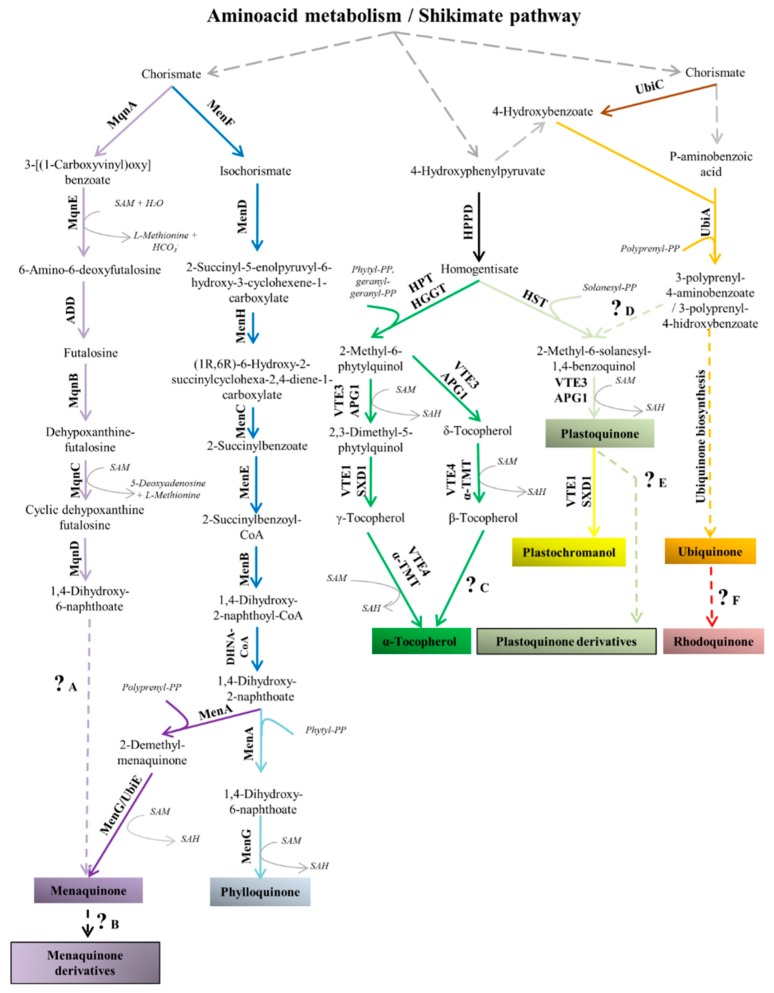
Figure 5.
Prenylquinone biosynthesis pathways [45]. Some prenylquinones shown have not been found in pathogenic protozoa, but all are cited in this review. The specific pathway of ubiquinone biosynthesis (orange arrows) and the formation of aromatic precursors from amino acid metabolism/shikimate pathways (gray arrows) are simplified because these processes are better described in other figures in this review. The incorporation of isoprene chains and use of S-adenosyl-L-methionine (SAM) by enzymes are indicated. The SAM cofactor is necessary for methylation reactions to produce S-adenosyl homocysteine or for other biochemical processes. Continuous arrows indicate a single enzymatic step, and discontinuous arrows indicate several enzymatic steps, processes that remain poorly understood or processes that are better shown in other figures in this review. The different pathways are distinguishable by the colors of the arrows: Light purple. MQ biosynthesis by the futalosine alternative pathway; Dark blue. Homologous enzymatic steps that are common for PK or MQ biosynthesis by the classic pathway; Dark purple. Specific enzymatic steps for MQ biosynthesis by the classic pathway; Light blue. Enzymatic steps that are specific for PK biosynthesis; Black. Biosynthesis of homogentisate from 4-hydroxyphenylpyruvate; Dark green. Tocopherol biosynthesis [142]; Light green. Shared enzymatic steps for PQ and plastochromanol biosynthesis; Yellow. Plastochromanol biosynthesis [45]; Brown. 4-Hydroxybenzoate biosynthesis from chorismate; Orange. Ubiquinone biosynthesis; Red. Possible rhodoquinone biosynthesis pathway in Rhodospirillum rubrum [143]. The figure also indicates processes that remain poorly understood. ?A. The final steps of the futalosine alternative pathway [144]; ?B. Biosynthesis of MQ derivatives such as ChQ or sulfated MQs (among other examples) [45]; ?C. α-Tocopherol biosynthesis from β-tocopherol; ?D. 2-Methyl-6-solanesyl-1,4-benzoquinol biosynthesis from 4-hydroxy-3-polyprenylbenzoate in cyanobacteria [145]; ?E. The formation of some PQ derivatives (e.g., PQ-B, PQ-C) is controversial in the literature [45]; ?F. Rhodoquinone biosynthesis requires ubiquinone in Rhodospirillum rubrum [143], but the process remains poorly understood. Next to each arrow, the corresponding enzyme is cited. For biosynthesis of tocopherol, plastochromanol and PQ, different enzymes can perform the same enzymatic step depending on the organism [141]. Enzymes: MqnA (chorismate dehydratase, EC:4.2.1.151). MqnE. (aminodeoxyfutalosine synthase, EC:2.5.1.120). ADD (aminodeoxyfutalosine deaminase, EC:3.5.4.40). MqnB (futalosine hydrolase, EC:3.2.2.26). MqnC (cyclic dehypoxanthinyl futalosine synthase, EC:1.21.98.1). MqnD (EC 1.14.). MenF (EC 5.4.4.2). MenD (EC 2.2.1.9). MenH (EC 4.2.99.20). MenC (EC 4.2.1.113). MenE (EC 6.2.1.26). MenB (EC 4.1.3.36). DHNA-CoA (EC 3.1.2.28). MenA (EC 2.5.1.74). MenG/UbiE (demethylmenaquinone methyltransferase/2-methoxy-6-polyprenyl-1,4-benzoquinol methylase; EC:2.1.1.163/EC:2.1.1.201, respectively). HPPD (EC:1.13.11.27). HPT (homogentisate phytyltransferase/homogentisate geranylgeranyltransferase; EC:2.5.1.115/EC:2.5.1.116, respectively). VTE3/APG1 (EC 2.1.1.295). VTE1/DXD1 (EC 5.5.1.24). VTE4/α-TMT (EC 2.1.1.95). HST (EC 2.5.1.117). UbiC (EC 4.1.3.40). UbiA (EC 2.5.1.39). Based on information from the cited references in the text and the Kyoto Encyclopedia of Genes and Genomes database [87].