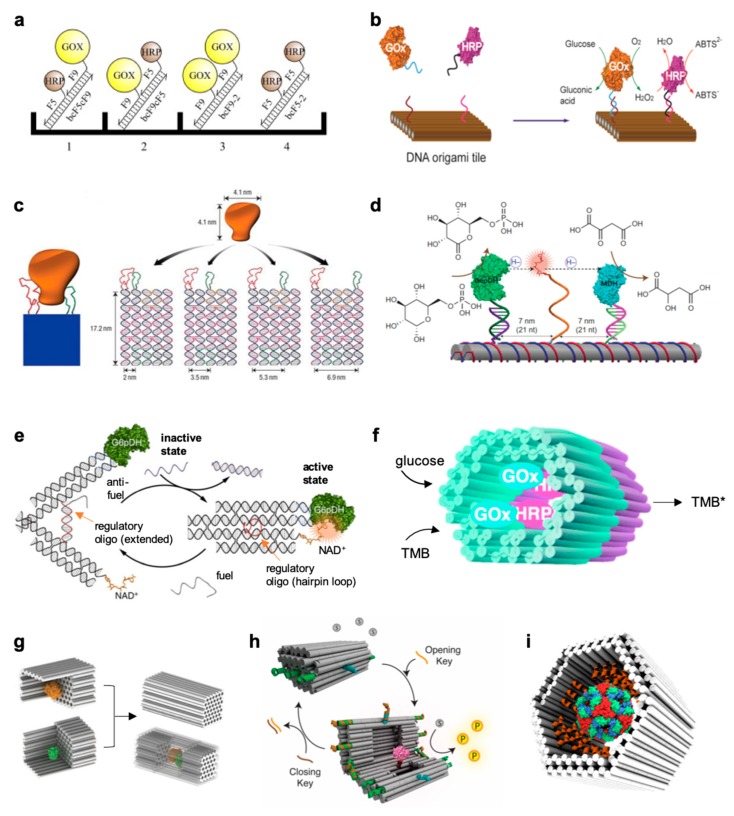
Figure 4.
DNA-scaffolded enzymes in different geometric configurations. (a) Four different complexes were assembled by DNA-directed immobilization on a microplate surface, with different arrangements of GOx and HRP in homologous and heterologous pairs [66]. (b) Coassembly of GOx and HRP enzymes on a two-dimensional DNA origami structure with control over inter-enzyme distances [71]. (c) Schematic showing a rigid DNA tile (blue) that can spatially separate two ligands (red and green) at a controlled distance, with each ligand attached to a different part of the target molecule (orange) for bivalent binding [72]. (d) Schematic of the nanostructured complex consisting of glucose-6-phosphate dehydrogenase (G6pDH) and a malic dehydrogenase (MDH) organized on a DNA DX tile [73]. The NAD-modified single-stranded poly(T)20 is positioned halfway between the two enzymes, facilitating the transfer of hydrides. (e) Schematic illustration of a DNA tweezer-regulated enzyme nanoreactor [74]: a regulatory oligomer (shown in red) is designed to adopt a hairpin structure that holds the two arms of the tweezers close together, thus bringing the dehydrogenase enzyme (G6pDH) and its cofactor (NAD+) in close proximity (active state). The addition of a fuel strand—complementary to the regulatory loop—results in the formation of a DNA double helix between the tweezer arms that separate the enzyme/cofactor pair (inactive state). (f) Two separately fabricated origami units are equipped with biotinylated glucose oxidase (GOx, cyan) and horseradish peroxidase (HRP, purple), respectively, through biotin–avidin interaction [75]. The units are linked together via base-pairing, resulting in a nanoreactor that is able to perform an enzyme cascade reaction. 3,3′,5,5′-tetramethylbenzidine (TMB) is oxidized into TMB diimine (TMB*) in the presence of glucose and oxygen. (g) Schematic representations of the assembly of a DNA nanocage encapsulating a pair of GOx (orange) and HRP (green) enzymes [76]. Individual enzymes were first attached to half cages, followed by the addition of linker strands to combine the two halves into a full cage. (h) Graphic representation of the DNA vault [77], a 3D dynamic DNA nanocontainer, which encapsulates and fully encloses a protease molecule (shown in pink) in its inner cavity, thus shielding it from its substrate (gray circles) that is free in solution, and preventing formation of the product (yellow circles). The opening and closing mechanisms are triggered by the addition of specific DNA strands (in orange and brown, respectively), which operate on the lock strands of the vault (green and blue). (i) A hollow DNA nanocontainer has been modified in its inner cavity with peptide recognition motifs (orange helices) pre-oriented toward the binding sites exposed on the surface of the DegP protease (multicolor sphere in the middle of the container), thus leading to selective encapsulation of a single protein molecule as a consequence of multivalent binding affinity and geometric matching of the two species [47]. Adapted with permission from References [47,66,71] and [72,73,74,75,76,77] © 2017 and 2016 Macmillan Publishers Ltd and 2015 Royal Society of Chemistry.