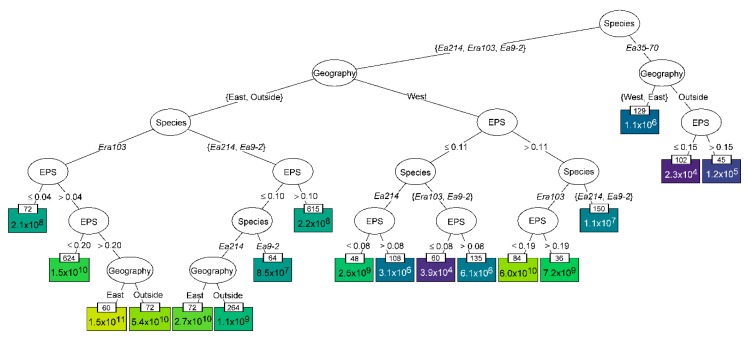
Figure 5.
Conditional inference tree explaining the effect of the phage species (Species), the global location of host isolation (Geography), the relative amylovoran of the host (EPS) and genus of host isolation (Genus) on the final genomic titre of a phage. Each node is continually split based on the most significant factor. In each white oval, the deciding factor of each split is given, and the specific determinants of splitting are shown below over the branch lines. In each coloured box is the mean genomic titre (genomes/mL) for each terminal node, with the number of observations given in the small white boxes above. This was a naïve analysis with no assumptions used to influence the splitting. Full species names are truncated, numbers are rounded, and p-values (all p < 0.001) are removed for simplicity. E. pyrifoliae and E. amylovora isolated from Rubus sp. sources were excluded from this analysis.