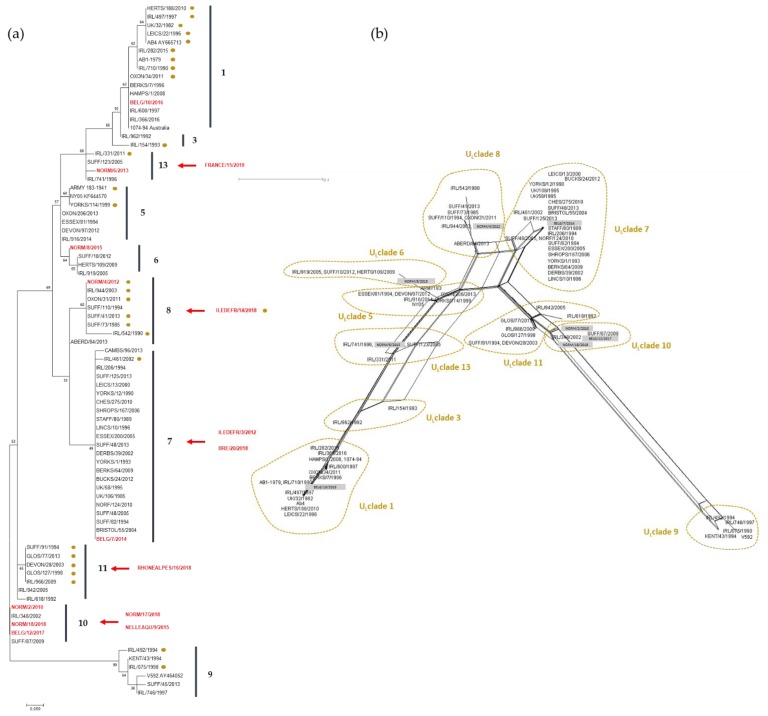
Figure 4.
(a) Maximum Likelihood phylogenic tree based on Jones Taylor Thornton model built with MLST sequences including 8 French and Belgian strains (in red), 66 EHV-1 UK strains [14], and 22 EHV-1 Irish strains [26]. Dots indicate D752 strains (Neuropathogenic type). Strains with partial concatenated amino acid sequence were not included in the Maximum Likelihood analysis but their supposed position in the tree is indicated with red arrows according to their clade identification (see Table 6). UL Clades [14] are numbered and represented with vertical lines. Boostrap values after 1000 replication are indicated at major nodes. (b) Network built using Neighbour-Net method with the same sequences as the tree. Only 37 amino acid sequences were included in the network, and French and Belgian strains are represented in grey boxes.