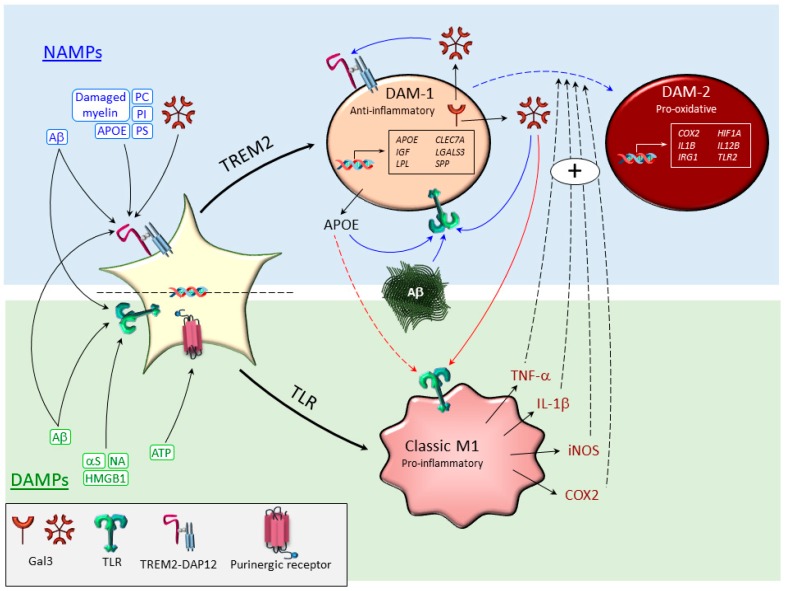
Figure 1.
Potential cross-interactions between different disease-associated microglia polarization subtypes. It is well established that microglia sense the disease environment through different pattern recognition receptors (PRRs). Two illustrative examples are toll-like receptors (TLRs) and triggering receptors expressed on myeloid cells-2 (TREM2). Specific ligands of PRRs are different danger-associated molecular patterns (DAMPs), including aggregated proteins (amyloid β, Aβ, and α-synuclein, αS); high-mobility group box protein 1 (HMGB1); nucleic acids (NA); and ATP. From these, Aβ and HMGB1 are believed to activate TLRs, thus driving microglia to a M1-proinflammatory phenotype, which is highly pro-oxidant. More recently, the term neurodegeneration-associated molecular patterns (NAMPs) has been introduced to highlight endogenous disease-associated ligands of TREM2. Examples of NAMPS include phosphatdyil serine (PS), present in apoptotic cells and glycolipids sphingomyelin and sulfatide derived from damaged myelin; Aβ; several lipoproteins like apolipoprotein E (APOE); and negatively charged phospholipids like phosphatidylinositol (PI) and phosphatidylcholine (PS). TREM2 signaling is suggested to drive the disease-associated microglia (DAM) phenotype, leading to downregulation of microglia homeostatic genes (not shown) and strong upregulation of DAM genes, including Apoe, Lgals3 (galectin-3; GAL3), Clec7a, etc., and thus, driving microglia to an anti-inflammatory (DAM-1) phenotype. From these, APOE and GAL3 can be released by reactive microglia and govern microglia immune responses. The possibility exists that GAL3 and APOE, along with other endogenous ligands like Aβ, drive TLR-associated signaling (solid blue arrows) further in either pro-oxidative DAM phenotypes (DAM-2; dashed blue arrow) or classic M1 proinflammatory microglias (red arrows). In addition, different classical microglia proinflammatory mediators like tumor necrosis factor (TNF)-α, interleukin (IL)-1β, inducible nitric oxide synthase (iNOS), and cyclooxygenase-2 (COX2) may affect the DAM phenotype, which may thus evolve into a pro-oxidative DAM phenotype.