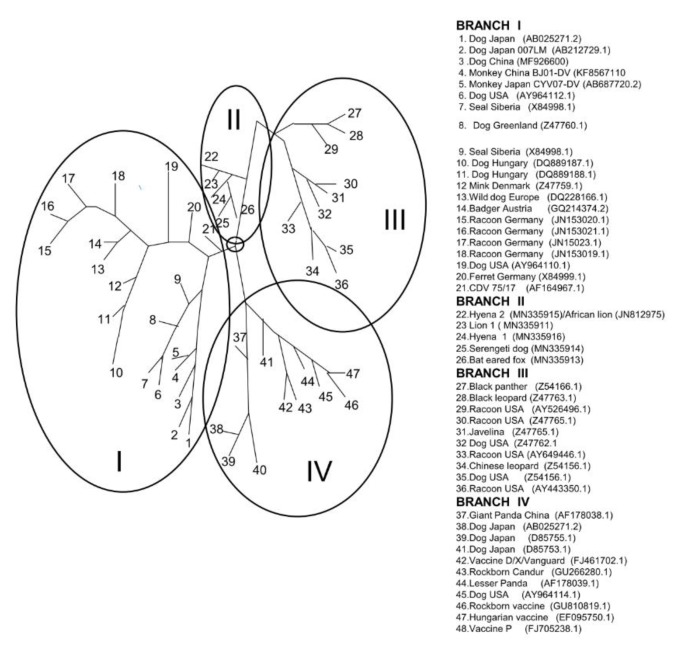
Figure 2.
Phylogenetic relationship of CDV strains from the 1993/1994 Serengeti epidemic to related viruses based on H gene sequences. Circles represent branches (I, II, III and IV) from a common node (black circle). Accession numbers are given for isolates in the figure key. Associated references where available for viruses as numbered in the key: 1, 37. Mochizuki et. al. [72]; 2. Lan et al. [73]; 3. Qiu et al [56]; 4. Feng et al. [71]; 5. Sakai et al. [57]; 6, 8, 12, 29, 30, 31, 34. Bolt et al. [74]; 7. 20. Mamaev et al. [5]; 8, 9, 10. 11 Demeter et al. [75]; 12. Martella et al. [62]; 14,15,16,17 18. Nikolin et al. [76]; 19, 43 Pardu et al. [66]; 21, Weiderkehr et al (unpublished); 22(Hyena)-24 Current report; 22 (lion). Nikolin et al. [52], 33. Harder et al. [77]; 28, 32, 35. Lednicky et al. [78]; 38, 39. Iwatsuki et al. [79]; 40. Woma et al. [64]; 41, 44. Martella et al. [80] 29: 1222–1227; 46. Chulakasian et al. [81]. Unrooted neighbor-joining phylogenetic trees were constructed by using the MegAlign version 7.1 package (DNASTAR, www.dnastar.com) with the ClustalW method (www.clustal.org).