Abstract
Emerging studies indicated that lncRNA is one crucial regulator in development and tumorigenesis. We firstly showed that LINC00994 was overexpressed in GC cells compare with GES-1. Compared to adjacent samples, the LINC00994 expression was increased in GC tissues by qRT-PCR assay. Furthermore, Elevated expression of LINC00994 induced GC cell invasion, proliferation and cycle. Luciferase reporter analysis indicated that ectopic miR-765-3p expression suppressed wild type LINC00994 luciferase activity, but not mutant LINC00994 in GC cells. Elevated expression of LINC00994 inhibited the miR-765-3p expression in GC cells. Compared to adjacent samples, the miR-765-3p expression was decreased in GC tissues by qRT-PCR assay. LINC00994 induced GC cell invasion and growth via modulating miR-765-3p. Thus, our data suggested an oncogenic role of LINC00994 in development of GC.
Keywords: LINC00994, miR-765-3p, gastric cancer, lncRNAs
Introduction
Gastric cancer is the 4th most common tumor and the 2nd leading cause of tumor-associated death worldwide [1-4]. The pathogenesis and mechanism of gastric cancer is complex and is related with several factors [5-7]. Despite achieving advances in these novel management strategies such as surgical techniques, chemotherapy and radiotherapy methods, the five year survival rate of this disease is still discontented [8-11]. Most gastric cancer patients were diagnosed at the advanced stage according to asymptomatic presentation at the early stage [12-14]. Therefore, it is useful to study molecular mechanisms and identify novel biomarkers and therapeutic targets for gastric cancer.
Long noncoding RNAs (lncRNAs) is one group of noncoding RNAs that exceed 200 nucleotides (nts) in length with no or limited protein coding capacity [15-19]. Growing studies indicated that lncRNAs participated in several cell biological processes such as stem cell pluripotency reprogramming, migration, invasion, proliferation and apotosis [20-23]. Several lncRNAs were found to be deregulated in a variety of tumors including lung carcinoma, hepatocellular carcinoma, bladder cancer, osteosarcoma and also gastric cancer [21,24-28]. Recently, one new lncRNA long intergenic non-protein coding RNA 994 (LINC00994) was identified and was found to be played important roles in the progression of some diseases such as type 2 diabetes (T2DM) and pancreatic cancer [29,30]. Mansoori and their colleagues reported that LINC00994 expression level was downregulated in the T2DM cases compared to control participants [29]. Zhu and their colleagues showed that LINC00994 silencing suppressed pancreatic tumor cell proliferation, invasion and migration and induced cell cycle arrest at the G1 stage and apoptosis [30]. However, the regulatory function of LINC00994 in the gastric cancer has not been investigated.
In this study, we firstly studied that LINC00994 was overexpressed in GC cells (AGS, SGC-7901, MGC8-03, and HGC-27) compare with GES-1. Compared to adjacent samples, the LINC00994 expression was increased in GC tissues by qRT-PCR assay. Elevated expression of LINC00994 induced GC cell invasion, proliferation and cycle.
Materials and methods
Tissues
Surgically resected gastric tumor and their adjacent samples were obtained from gastric caner cases that underwent surgery at our department. Surgically collected samples were immediately frozen in the liquid nitrogen until use. All patients were signed the informed consent and our research was performed following to Helsinki principles Declaration and ethics committee of Ningxia Medical University.
Cell lines cultured and transfection
Four human gastric cancer cell lines (AGS, SGC-7901, MGC8-03, and HGC-27) and gastric epithelial cell line (GES-1) were buying from cell bank of Chinese Academy of Sciences (Shanghai, China). These cell lines were plated in the medium of RPMI-1640 supplemented with FBS (fetal bovine serum) and antibiotics. pcDNA-LINC00994 and its control vector, miR-765-3p mimic, inhibitor and control were obtained from the RiboBi company (Guangzhou, China) and then were transfected to cells by using Lipofectamine 2000 kit (Invitrogen, Carlsbad, USA).
RNA extraction and real-time quantitative PCR
Total RNA was distilled from cell or sample with Rneasy Mini (Qiagen, Germany) following to protocol of manufacture. LINC00994 and miR-765-3p expression were tested by SYBR (Takara, China) in a PCR system of Bio-Rad (VisonBio Scientific). Data were normalized to U6 or GAPDH. Primer sequences were indicated as following: The 2-DDCT way was exploited for PCR data analysis. Primer information was shown as following: miR-765-3p, sense: 5’-ATCAGTGGAGGAGAAGGAAGG-3’ and antisense: 5’-GTGCAGGGTCCGAGGTATTC-3’; LINC00994, sense: 5’-TCAAGGAGCTGGGATGGA-3’ and antisense: 5’-CGAATTGCTGGGAAGAGG-3’.
Cell proliferation, cell cycle and migration assays
Cell growth was carried out using CCK-8 aasay (Dojin, Japan) at different time point. Cells were plated in the 96-well and CCK-8 fluid was added into each plate well. After 2 hours incubation, the absorbance at 450 nm was checked at Multiplate Reader (Thermo Scientific). Cell was fixed by ice-cold 70% ethanol overnight and then stained with PI (propidium iodide, Beyotime) supplement with RNase A in dark environment. Cell cycle was carried out on flow cytometer (Biosciences, USA). Transwell analysis was maken to evaluate cell invasion. Cells were keplt in upper chamber and FBS was added to lower chamber. After 48 h, invasive cells were fixed, stained and counted.
Luciferase reporter assay
miR-765-3p binding sites on LINC00994 were seeked from starBase (http://starbase.sysu.edu.cn/index.php). Wild-type (Wt) binding type and mutant binding type of LINC00994 were synthesized and produced LINC00994-wt and LINC00994-mut. Cells were transfected with miR-765-3p mimic and scramble, LINC00994-wt or LINC00994-mut. Luciferase activity was detected by Luciferase Reporter analysis kit (Promega, Madison, USA) after transfection with 48 h.
Statistical analysis
Data were shown as the mean value and the standard deviation (SD) was presented the error bars. The statistical comparisons between two groups were analyzed by student’s t test and data between more than two groups were shown with one-way ANOVA. P value <0.05 was indicated as significant.
Results
LINC00994 was overexpressed in GC cells
LINC00994 expression was examined by qRT-PCR in the GC cell lines. As indicated in Figure 1A, LINC00994 was overexpressed in GC cells (AGS, SGC-7901, MGC8-03, and HGC-27) compare with GES-1. In line with this, the expression of LINC00994 was upregulated in GC cells (AGS, SGC-7901, MGC8-03, and HGC-27) compare to GES-1 by PCR (Figure 1B).
Figure 1.
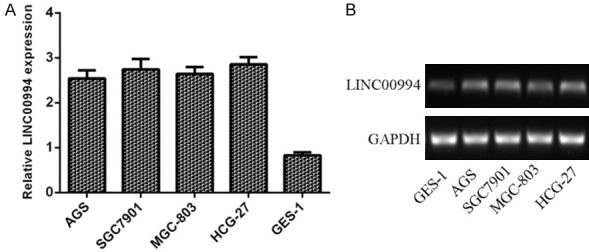
LINC00994 was overexpressed in GC cells. A. The expression level of LINC00994 was analyzed in GC cells (AGS, SGC-7901, MGC8-03, and HGC-27) and gastric epithelial cell line (GES-1) by using qRT-PCR analysis. B. The expression of LINC00994 in GC cells (AGS, SGC-7901, MGC8-03, and HGC-27) and GES-1 was measured by PCR.
LINC00994 was upregulated in GC samples
Compared to adjacent samples, the LINC00994 expression was increased in GC tissues by qRT-PCR assay (Figure 2A). Furthermore, LINC00994 was overexpression in 31 GC cases (31/40; 77.5%) compared to adjacent specimens (Figure 2B).
Figure 2.
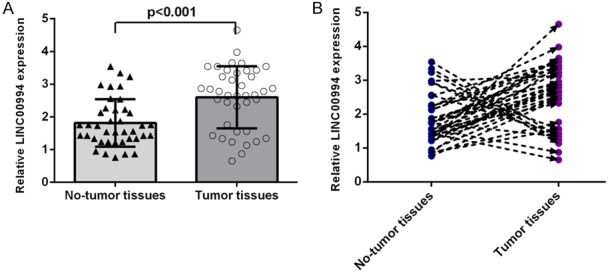
LINC00994 was upregulated in GC samples. A. Compared to adjacent samples, the LINC00994 expression was increased in GC tissues by qRT-PCR assay. B. LINC00994 was overexpression in 31 GC cases (31/40; 77.5%) compared to adjacent specimens.
Elevated expression of LINC00994 induced GC cell proliferation and cycle
Then, we discussed the cell function of LINC00994 in GC cell, we established a LINC00994 overexpression plasmid. We verified that the LINC00994 expression was upregulated both in the SGC-7901 (Figure 3A) and HGC-27 cells (Figure 3B). CCK-8 analysis data indicated that LINC00994 overexpression promoted SGC-7901 cell growth (Figure 3C) and elevated expression of LINC00994 increased HGC-27 cell proliferation (Figure 3D). Moreover, ectopic LINC00994 expression enhanced the cell cycle both in the SGC-7901 cell (Figure 3E) and HGC-27 cells (Figure 3F).
Figure 3.
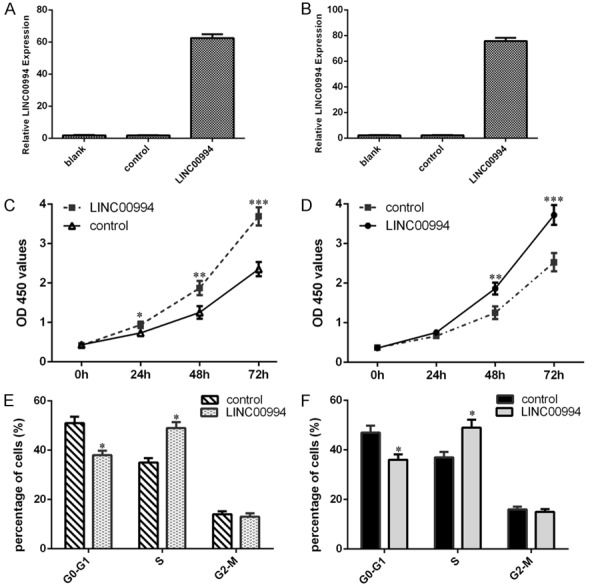
Elevated expression of LINC00994 induced GC cell proliferation and cycle. A. LINC00994 expression was upregulated in the SGC-7901 cell after transfection with pcDNA-LINC00994. B. The expression level of LINC00994 was measured by qRT-PCR analysis in the HGC-27 cell. C. LINC00994 overexpression promoted SGC-7901 cell growth by CCK-8 analysis. D. Elevated expression of LINC00994 increased HGC-27 cell proliferation. E. Ectopic LINC00994 expression enhanced the cell cycle both in the SGC-7901 cell. F. Overexpression of LINC00994 promoted cell cycle both in the HGC-27 cell. *P<0.05, **P<0.01 and ***P<0.001.
Ectopic LINC00994 expression enhanced GC cell invasion
Furthermore, the function of LINC00994 overexpression on cell invasion was measured using invasion transwell analysis. It was shown that elevated expression of LINC00994 induced the SGC-7901 cell invasion (Figure 4A and 4B). Overexpression of LINC00994 increased cell invasion in the SGC-7901 cell (Figure 4C and 4D).
Figure 4.
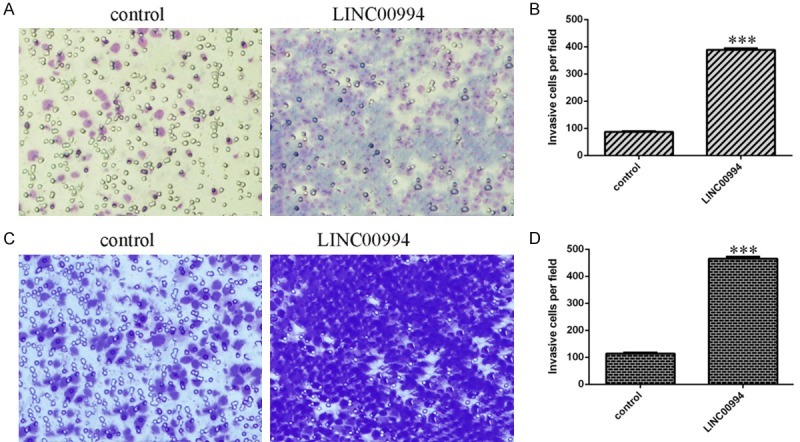
Ectopic LINC00994 expression enhanced GC cell invasion. A. Ectopic LINC00994 expression promoted SGC-7901 cell invasion. B. The invasive cells were shown. C. Overexpression of LINC00994 increased cell invasion in the SGC-7901 cell. D. The invasive cells were shown. ***P<0.001.
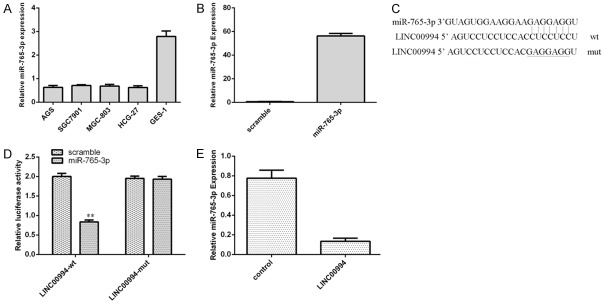
LINC00994 can sponge miR-765-3p expression in GC cell
As indicated in Figure 5A, miR-765-3p was downregulated in GC cells (AGS, SGC-7901, MGC8-03, and HGC-27) compare with GES-1. We verified that the miR-765-3p expression was upregulated both in the SGC-7901 cells (Figure 5B). Seeking from starBase (http://starbase.sysu.edu.cn/index.php), miR-765-3p harbored binding sites with LINC00994 (Figure 5C). Luciferase reporter analysis indicated that ectopic miR-765-3p expression suppressed wild type LINC00994 luciferase activity, but not mutant LINC00994 in SGC-7901 cells (Figure 5D). Elevated expression of LINC00994 inhibited the miR-765-3p expression in SGC-7901 cells (Figure 5E).
Figure 5.
LINC00994 can sponge miR-765-3p expression in GC cell. A. miR-765-3p was downregulated in GC cells (AGS, SGC-7901, MGC8-03, and HGC-27) compare with GES-1. B. miR-765-3p expression was upregulated both in the SGC-7901 cells after transfection with miR-765-3p mimic. C. Seeking from starBase (http://starbase.sysu.edu.cn/index.php), miR-765-3p harbored binding sites with LINC00994. D. Ectopic miR-765-3p expression suppressed wild type LINC00994 luciferase activity, but not mutant LINC00994 in SGC-7901 cells. E. Elevated expression of LINC00994 inhibited the miR-765-3p expression in SGC-7901 cells. **P<0.01.
miR-765-3p was downregulated in GC samples
Compared to adjacent samples, the miR-765-3p expression was decreased in GC tissues by qRT-PCR assay (Figure 6A). Furthermore, miR-765-3p was donwregulated in 29 GC cases (29/40; 72.5 %) compared to adjacent specimens (Figure 6B). Pearson’s correlation was done to measure correlation between the expression of miR-765-3p and LINC00994. Data indicated that LINC00994 level was inversely correlated with miR-765-3p in cases with GC (Figure 6C).
Figure 6.
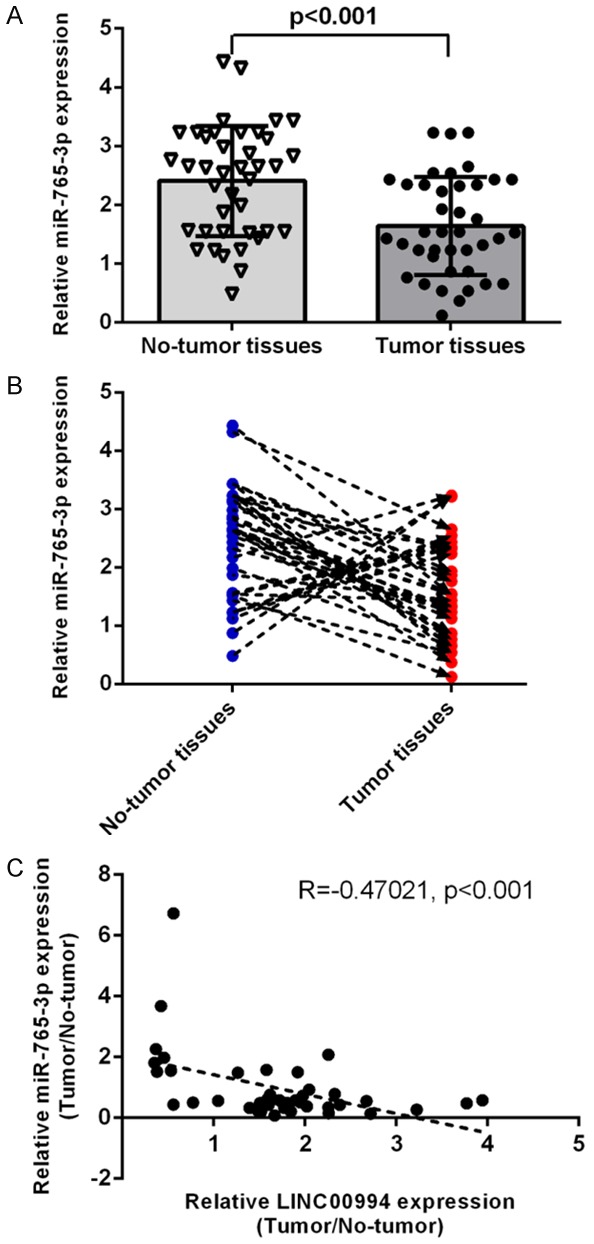
MiR-765-3p was downregulated in GC samples. A. Compared to adjacent samples, the miR-765-3p expression was decreased in GC tissues by qRT-PCR assay. B. miR-765-3p was donwregulated in 29 GC cases (29/40; 72.5%) compared to adjacent specimens. C. LINC00994 level was inversely correlated with miR-765-3p in cases with GC.
LINC00994 induced GC cell invasion and growth via modulating miR-765-3p
We performed rescue analysis to study role of LINC00994/miR-765-3p in modulating GC cell invasion and proliferation. As shown in Figure 7A, cell growth enhanced by transfection with pcDNA-LINC00994 was partly rescued by miR-765-3p mimic introduction. In addition, we found that miR-765-3p overexpression decreased the cell cycle in the LINC00994-overexpressing SGC-7901 cells (Figure 7B). Additionally, our data indicated that ectopic miR-765-3p expression suppressed cell invasion in the LINC00994-overexpressing SGC-7901 cells (Figure 7C and 7D).
Figure 7.
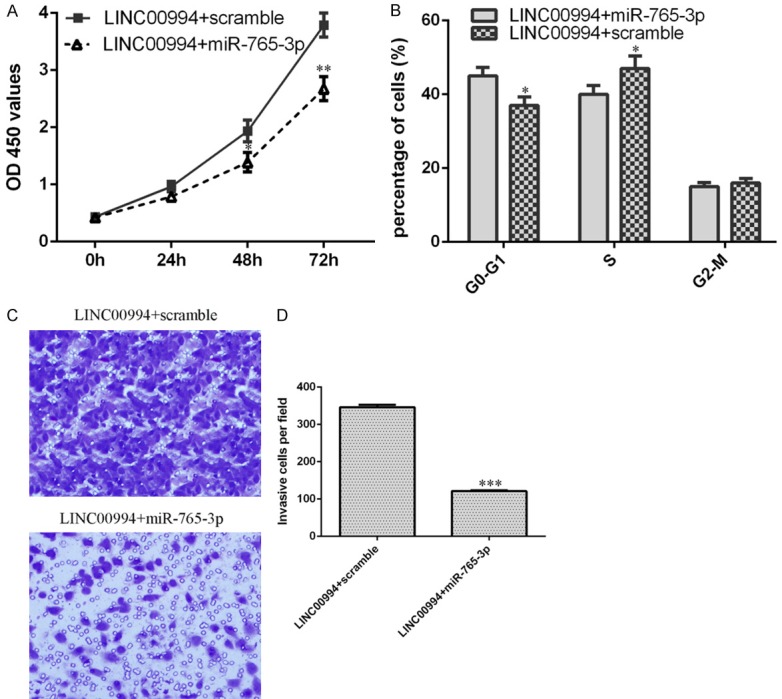
LINC00994 induced GC cell invasion and growth via modulating miR-765-3p. A. The cell proliferation in differenct group was detected by CCK-8 assay. B. miR-765-3p overexpression decreased the cell cycle in the LINC00994-overexpressing SGC-7901 cells. C. Ectopic miR-765-3p expression suppressed cell invasion in the LINC00994-overexpressing SGC-7901 cells. D. The invasive cells were shown. *P<0.05, **P<0.01 and ***P<0.001.
Discussion
LncRNAs are found to act as critical regulator in cell processes through controling gene expression at epigenetic, post-transcriptional and transcriptional level [31-34]. In this research, we firstly showed that LINC00994 was overexpressed in GC cells (AGS, SGC-7901, MGC8-03, and HGC-27) compare with GES-1. Compared to adjacent samples, the LINC00994 expression was increased in GC tissues by qRT-PCR assay. Furthermore, LINC00994 was overexpression in 31 GC cases (31/40; 77.5%) compared to adjacent specimens. Elevated expression of LINC00994 induced GC cell invasion, proliferation and cycle. Luciferase reporter analysis indicated that ectopic miR-765-3p expression suppressed wild type LINC00994 luciferase activity, but not mutant LINC00994 in GC cells. Elevated expression of LINC00994 inhibited the miR-765-3p expression in GC cells. Compared to adjacent samples, the miR-765-3p expression was decreased in GC tissues by qRT-PCR assay. Additional, miR-765-3p was donwregulated in 29 GC cases (29/40; 72.5%) compared to adjacent specimens. LINC00994 induced GC cell invasion and growth via modulating miR-765-3p. Thus, our data suggested an oncogenic role of LINC00994 in development of GC.
LINC00994 was found to be downregulated in type 2 diabetes in the Iranian cohort [29]. Recently, Zhu et al [30]. showed that LINC00994 expression was overexpressed in pancreatic cancer samples and knockdown LINC00994 expression suppressed cell migration, growth and invasion and induced cell apoptosis and cycle arrest at G1 stage. Nevertheless, function role of LINC00994 in GC remains almost unknown. We firstly detected the LINC00994 expression in GC cells (AGS, SGC-7901, MGC8-03, and HGC-27) and GES-1. Our data indicated that LINC00994 was overexpressed in GC cells compare with GES-1. Compared to adjacent samples, the LINC00994 expression was increased in GC tissues by qRT-PCR assay. Furthermore, LINC00994 was overexpression in 31 GC cases (31/40; 77.5%) compared to adjacent specimens. Ectopic expression of LINC00994 induced GC cell invasion, proliferation and cycle.
Growing references suggested that LncRNAs play as miRNA sponges to inhibit the modulatory function of miRNAs, which cause post-transcriptional regulation on potential mRNAs [35,36]. For instance, Wang et al [37]. demonstrated that UCA1 induced migration and growth and suppressed cell apoptosis via sponging miRNAs. Guo et al [38]. indicated that TUBA4B decreased GC cell progression via regulating miR-216b/a and miR-214. Dong et al [39]. found that GAS5 overexpression decreased development and tumorigenesis of GC via sponging miR-106a-5p. Huang et al [40]. demonstrated that TRPM2-AS induced GC progression through modulating miR-195. Zhu et al [30]. showed that LINC00994 inhibition inhibited pancreatic tumor cell progression via regulating miR-765-3p. Seeking from starBase (http://starbase.sysu.edu.cn/index.php), miR-765-3p harbored binding sites with LINC00994. Luciferase reporter analysis indicated that ectopic miR-765-3p expression suppressed wild type LINC00994 luciferase activity, but not mutant LINC00994 in SGC-7901 cells. Elevated expression of LINC00994 inhibited the miR-765-3p expression in SGC-7901 cells. Compared to adjacent samples, the miR-765-3p expression was decreased in GC tissues. Pearson’s correlation data indicated that LINC00994 level was inversely correlated with miR-765-3p in cases with GC.
Our data suggested that LINC00994 expression was overexpressed in GC cells and samples and ectopic LINC00994 expression promoted GC cell cycle, invasion and proliferation through modulating miR-765-3p. Thus, our data suggested an oncogenic role of LINC00994 in development of GC.
Acknowledgements
This study was supported by Ningxia high school Top Discipline construction (Traditional Chinese Medicine Discipline, No. NXYLXK2017A06) funded project.
Disclosure of conflict of interest
None.
References
- 1.Zou J, Xu Y. MicroRNA-140 inhibits cell proliferation in gastric cancer cell line HGC-27 by suppressing SOX4. Med Sci Monit. 2016;22:2243–2252. doi: 10.12659/MSM.896633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang M, Dong BB, Lu M, Zheng MJ, Chen H, Ding JZ, Xu AM, Xu YH. miR-429 functions as a tumor suppressor by targeting FSCN1 in gastric cancer cells. Onco Targets Ther. 2016;9:1123–1133. doi: 10.2147/OTT.S91879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu M, Zhou X, Du Y, Huang Z, Zhu J, Xu J, Cheng G, Shu Y, Liu P, Zhu W, Wang T. miR-20a induces cisplatin resistance of a human gastric cancer cell line via targeting CYLD. Mol Med Rep. 2016;14:1742–1750. doi: 10.3892/mmr.2016.5413. [DOI] [PubMed] [Google Scholar]
- 4.Zhuang K, Han K, Tang H, Yin X, Zhang J, Zhang X, Zhang L. Up-regulation of plasma miR-23b is associated with poor prognosis of gastric cancer. Med Sci Monit. 2016;22:356–361. doi: 10.12659/MSM.895428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Lei H, Luo M, Wang Y, Dong L, Ma Y, Liu C, Song W, Wang F, Zhang J, Shen J, Yu J. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18:43–54. doi: 10.1007/s10120-014-0340-8. [DOI] [PubMed] [Google Scholar]
- 6.Yu B, Lv X, Su L, Li J, Yu Y, Gu Q, Yan M, Zhu Z, Liu B. MiR-148a functions as a tumor suppressor by targeting CCK-BR via inactivating STAT3 and Akt in human gastric cancer. PLoS One. 2016;11:e0158961. doi: 10.1371/journal.pone.0158961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X, Zhang Y, Liu Z, Zhang X, Jia J. miRNA-532-5p functions as an oncogenic microRNA in human gastric cancer by directly targeting RUNX3. J Cell Mol Med. 2016;20:95–103. doi: 10.1111/jcmm.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie SS, Jin J, Xu X, Zhuo W, Zhou TH. Emerging roles of non-coding RNAs in gastric cancer: pathogenesis and clinical implications. World J Gastroenterol. 2016;22:1213–1223. doi: 10.3748/wjg.v22.i3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C, Zheng X, Li X, Fesler A, Hu W, Chen L, Xu B, Wang Q, Tong A, Burke S, Ju J, Jiang J. Reduction of gastric cancer proliferation and invasion by miR-15a mediated suppression of Bmi-1 translation. Oncotarget. 2016;7:14522–14536. doi: 10.18632/oncotarget.7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Zeng J, Pan J, Geng X, Li L, Wu J, Song P, Wang Y, Liu J, Wang L. MiR-320a inhibits gastric carcinoma by targeting activity in the FoxM1-P27KIP1 axis. Oncotarget. 2016;7:29275–29286. doi: 10.18632/oncotarget.8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LL, Zhang XH, Zhang X, Chu JK. MiR-30a increases cisplatin sensitivity of gastric cancer cells through suppressing epithelial-to-mesenchymal transition (EMT) Eur Rev Med Pharmacol Sci. 2016;20:1733–1739. [PubMed] [Google Scholar]
- 12.Zhang Z, Dou M, Yao X, Tang H, Li Z, Zhao X. Potential biomarkers in diagnosis of human gastric cancer. Cancer Invest. 2016;34:115–122. doi: 10.3109/07357907.2015.1114122. [DOI] [PubMed] [Google Scholar]
- 13.Vidal AF, Cruz AM, Magalhaes L, Pereira AL, Anaissi AK, Alves NC, Albuquerque PJ, Burbano RM, Demachki S, Ribeiro-dos-Santos A. hsa-miR-29c and hsa-miR-135b differential expression as potential biomarker of gastric carcinogenesis. World J Gastroenterol. 2016;22:2060–2070. doi: 10.3748/wjg.v22.i6.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong D, Zhao L, He K, Sun H, Cai D, Ni L, Sun R, Chang S, Song T, Huang C. MECP2 promotes the growth of gastric cancer cells by suppressing miR-338-mediated antiproliferative effect. Oncotarget. 2016;7:34845–34859. doi: 10.18632/oncotarget.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, Li Z. Long non-coding RNA growth arrest-specific transcript 5 in tumor biology. Oncol Lett. 2015;10:1953–1958. doi: 10.3892/ol.2015.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Yu X, Shen J. Long non-coding RNAs: emerging players in osteosarcoma. Tumor Biol. 2016;37:2811–2816. doi: 10.1007/s13277-015-4749-4. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Li X, Chen X, Li S, Ho IHT, Liu X, Chan MTV, Wu WKK. Emerging roles of long non-coding RNAs in neuropathic pain. Cell Prolif. 2019;52:e12528. doi: 10.1111/cpr.12528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Li X, Chen C, Li S, Shen J, Tse G, Chan MTV, Wu WKK. Long non-coding RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2018;51:e12483. doi: 10.1111/cpr.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng J, Yi D, Liu Y, Wang M, Zhu Y, Shi H. Long nonding RNA UCA1 regulates neural stem cell differentiation by controlling miR-1/Hes1 expression. Am J Transl Res. 2017;9:3696–3704. [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C, Wu K, Wang S, Wei G. Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J Cell Biochem. 2018;119:5646–5656. doi: 10.1002/jcb.26743. [DOI] [PubMed] [Google Scholar]
- 21.Zhao J, Gao Z, Zhang C, Wu H, Gu R, Jiang R. Long non-coding RNA ASBEL promotes osteosarcoma cell proliferation, migration and invasion by regulating microRNA-21. J Cell Biochem. 2018;119:6461–6469. doi: 10.1002/jcb.26671. [DOI] [PubMed] [Google Scholar]
- 22.Zhang LM, Wang P, Liu XM, Zhang YJ. LncRNA SUMO1P3 drives colon cancer growth, metastasis and angiogenesis. Am J Transl Res. 2017;9:5461–5472. [PMC free article] [PubMed] [Google Scholar]
- 23.Ye K, Wang S, Zhang H, Han H, Ma B, Nan W. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J Cell Biochem. 2017;118:4772–4781. doi: 10.1002/jcb.26145. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Zhang F, Wang J, Hu L, Chen J, Xu G, Wang Y. LncRNA LINC01512 promotes the progression and enhances oncogenic ability of lung adenocarcinoma. J Cell Biochem. 2017;118:3102–3110. doi: 10.1002/jcb.26178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.She K, Yan H, Huang J, Zhou H, He J. miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer. Cell Prolif. 2018;51 doi: 10.1111/cpr.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He A, Liu Y, Chen Z, Li J, Chen M, Liu L, Liao X, Lv Z, Zhan Y, Zhuang C, Lin J, Huang W, Mei H. Over-expression of long noncoding RNA BANCR inhibits malignant phenotypes of human bladder cancer. J Exp Clin Cancer Res. 2016;35:125. doi: 10.1186/s13046-016-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen JF. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansoori Z, Ghaedi H, Sadatamini M, Vahabpour R, Rahimipour A, Shanaki M, Saeidi L, Kazerouni F. Downregulation of long non-coding RNAs LINC00523 and LINC00994 in type 2 diabetes in an Iranian cohort. Mol Biol Rep. 2018;45:1227–1233. doi: 10.1007/s11033-018-4276-7. [DOI] [PubMed] [Google Scholar]
- 30.Zhu X, Niu X, Ge C. Inhibition of LINC00994 represses malignant behaviors of pancreatic cancer cells: interacting with miR-765-3p/RUNX2 axis. Cancer Biol Ther. 2019;20:799–811. doi: 10.1080/15384047.2018.1564566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Lu X, Geng Z, Yang G, Shi Y. LncRNA PTCSC3/miR-574-5p governs cell proliferation and migration of papillary thyroid carcinoma via Wnt/-Catenin signaling. J Cell Biochem. 2017;118:4745–4752. doi: 10.1002/jcb.26142. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Lv G, Li J, Wang B, Zhang Q, Lu C. LncRNA-RP11-296A18.3/miR-138/HIF1A pathway regulates the proliferation ECM synthesis of human nucleus pulposus cells (HNPCs) J Cell Biochem. 2017;118:4862–4871. doi: 10.1002/jcb.26166. [DOI] [PubMed] [Google Scholar]
- 33.Wang G, Pan J, Zhang L, Wei Y, Wang C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS1. Cell Prolif. 2017;50 doi: 10.1111/cpr.12389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Ma X, Li Z, Li T, Zhu L, Li Z, Tian N. Long non-coding RNA HOTAIR enhances angiogenesis by induction of VEGFA expression in glioma cells and transmission to endothelial cells via glioma cell derived-extracellular vesicles. Am J Transl Res. 2017;9:5012–5021. [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, Shen W, Li X, Zhang L, Jin X. The lncRNA n340790 accelerates carcinogenesis of thyroid cancer by regulating miR-1254. Am J Transl Res. 2017;9:2181–2194. [PMC free article] [PubMed] [Google Scholar]
- 36.Huan J, Xing L, Lin Q, Xui H, Qin X. Long noncoding RNA CRNDE activates Wnt/beta-catenin signaling pathway through acting as a molecular sponge of microRNA-136 in human breast cancer. Am J Transl Res. 2017;9:1977–1989. [PMC free article] [PubMed] [Google Scholar]
- 37.Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q, Zhao G, Zhang ZZ. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18:115. doi: 10.1186/s12943-019-1032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo J, Li Y, Duan H, Yuan L. LncRNA TUBA4B functions as a competitive endogenous RNA to inhibit gastric cancer progression by elevating PTEN via sponging miR-214 and miR-216a/b. Cancer Cell Int. 2019;19:156. doi: 10.1186/s12935-019-0879-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Dong S, Zhang X, Liu D. Overexpression of long noncoding RNA GAS5 suppresses tumorigenesis and development of gastric cancer by sponging miR-106a-5p through the Akt/mTOR pathway. Biol Open. 2019;8 doi: 10.1242/bio.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang B, Chang C, Wang BL, Li H. ELK1-induced upregulation of lncRNA TRPM2-AS promotes tumor progression in gastric cancer by regulating miR-195/HMGA1 axis. J Cell Biochem. 2019;120:16921–16933. doi: 10.1002/jcb.28951. [DOI] [PubMed] [Google Scholar]