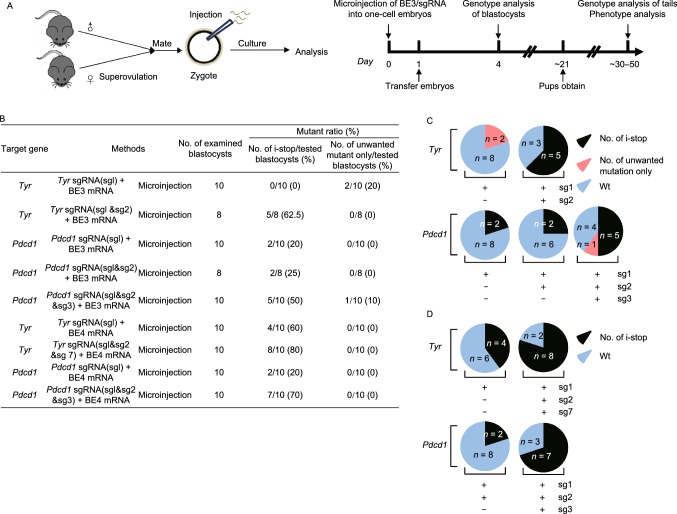
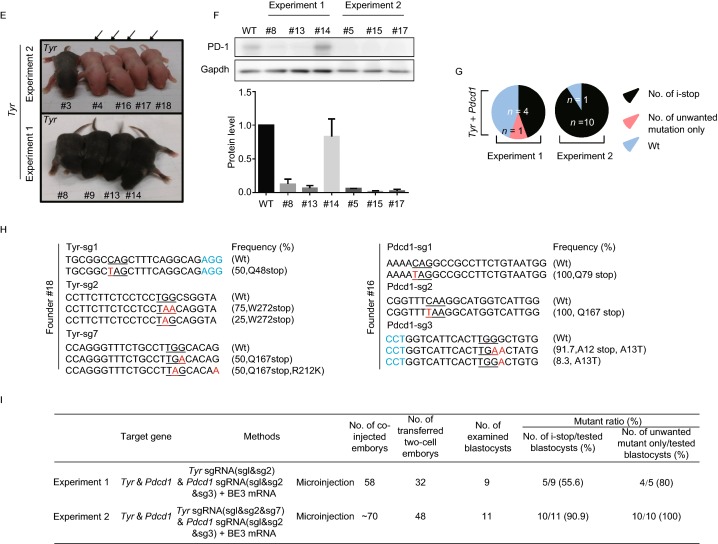
Figure 1.
Efficient C-to-T substitution at Tyr and Pdcd1 loci in mouse embryos and mutant mice. (A) Representative schematic and timeline of experimental design. After mating and superovulation of mice, sgRNA and BE3 mRNA or BE4max mRNA were co-injected into one-cell embryos, then editing efficiency were detected at blastocyst stage and founder mice. (B) Summary of embryo manipulation. (C) The percentage of different mutation types in mouse embryos by BE3-mediated base editing. Black represents percentage of blastocysts harboring generated stop codons; Red represents percentage of blastocysts harboring unwanted mutations only; Blue represents percentage of Wt (wild type) blastocysts. The number was indicated on the chart. (D) The percentage of different mutation types in mouse embryos by BE4max-mediated base editing. Black represents percentage of blastocysts harboring generated stop codons; Blue represents percentage of Wt (wild type) blastocysts. The number was indicated on the chart. (E) Tyr mutant newborn pups that developed after co-injecting the BE3 mRNA and sgRNA exhibited albino phenotype in their eyes and skin (black arrows, #4, #16, #17 and #18). (F) Representative results of phenotypes of mice from Pdcd1 targeting. Western blot (WB) showing that knockout of Pdcd1 leads to a decrease in PD-1 protein of #5, #8, #13, #15 and #17. (G) The percentage of different mutation types in pups. Black represents percentage of blastocysts harboring induced stop codons; Red represents percentage of blastocysts harboring unwanted mutations only; Blue represents percentage of wild type blastocysts. The number was indicated on the chart. (H) Representative alignments of modified sequences from newborn pups (#16 and #18) using microinjection of BE3 mRNA and sgRNAs into one-stage embryos. The PAM sequences and substitutions are highlighted in blue and red, respectively; The expectedly edited codons are underlined. (I) Summary of the numbers of embryos used and mutants targeting the Tyr and Pdcd1 sites