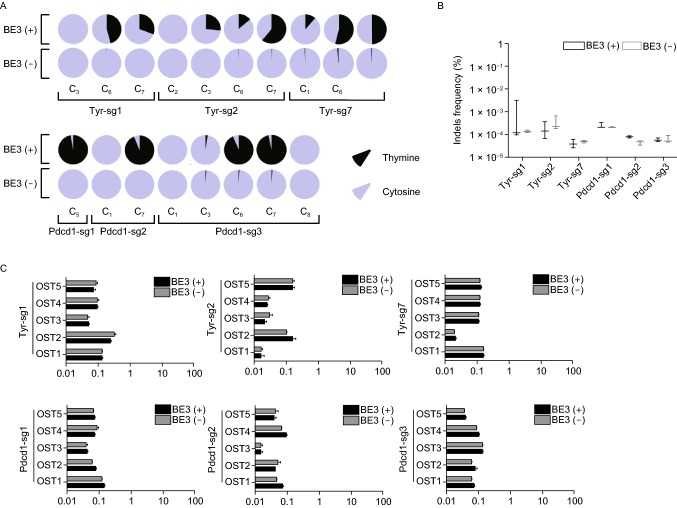
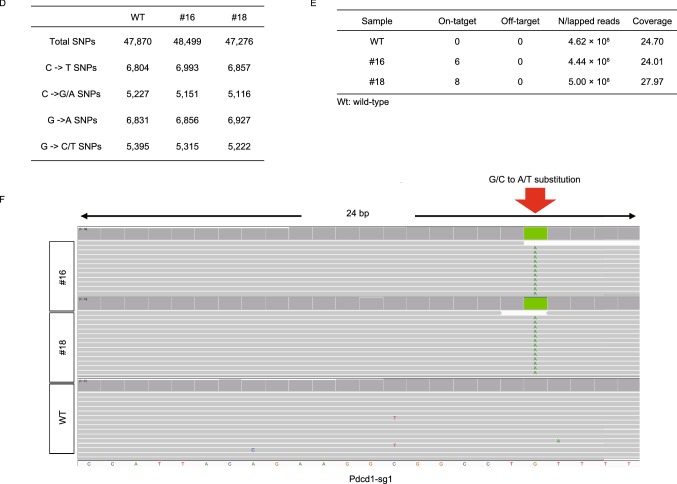
Figure 2.
Comprehensively on- and off-target analysis using targeted deep sequencing and WGS. (A) The editing frequency of thymine and cytosine were plotted. Black represents thymine and gray represents cytosine. The positions of edited Cs in the Tyr-sg1, Tyr-sg2, Tyr-sg7, Pdcd1-sg1, Pdcd1-sg2 and Pdcd1-sg3 target regions were indicated with the base distal from the PAM set as position 1. (B) The frequencies of desired C-to-T editing to unwanted indels. Statistical analyses show no significant differences between BE3 (+) (black) and BE3 (−) (gray) in indels. (C) Off-target analysis by PCR amplicon-based deep sequencing. The black column represents BE3 (+) and the gray column represents BE3 (−). (D) Summary of genome sequencing analysis. Two mutant mice (#16 and #18) and a wild-type mouse (WT) were sequenced separately using Illumina Novaseq. A total of 47,870, 48,499 and 47,276 SNPs were identified for Wt, #16 and #18, respectively. (E) Summary of on- and off-target analysis. (F) Confirmation of the on-target mutation by the analysis of whole-genome sequencing. Red arrow indicates the G/C to A/T substitution within 24 bp on-target sequence for Pdcd1-sg1