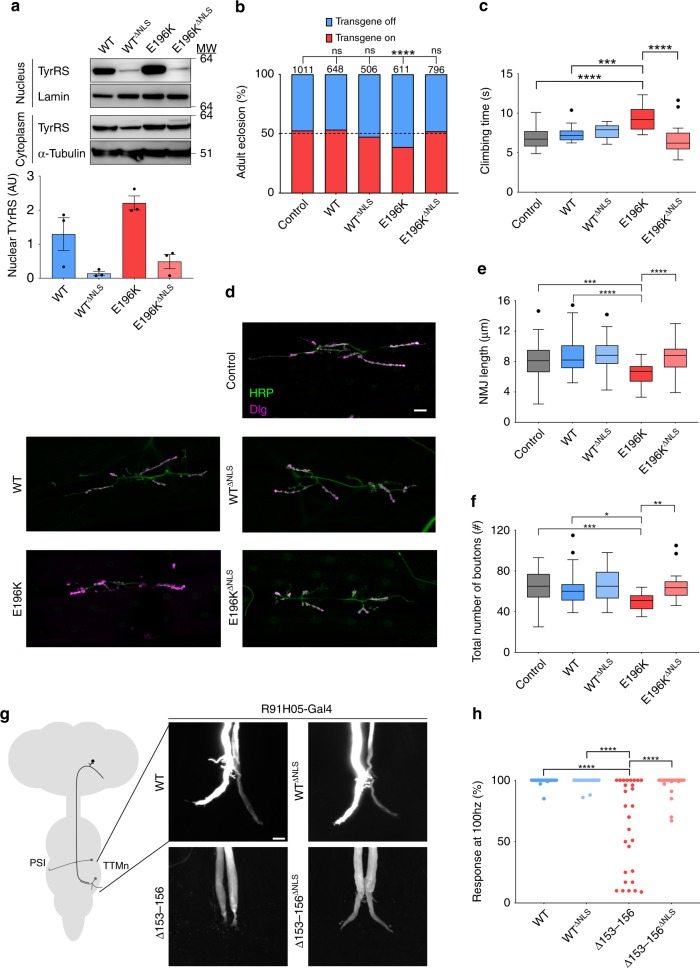
Fig. 5.
Genetic nuclear exclusion of mutant TyrRS fully prevents disease phenotypes in Drosophila (a) Subcellular localization of TyrRS in heads of flies pan-neuronally (nSyb-Gal4) expressing different TyrRS alleles. ΔNLS, disrupted NLS motif by mutating 242KKKLKK247 to 242NNKLNK247. α-Tubulin and Lamin served as cytoplasmic and nuclear markers, respectively. n = 3 independent biological replicates, bar charts presented as means ± s.e.m. b Eclosion rates of flies ubiquitously expressing (Act5C-Gal4) no transgene (Control) or different TyrRS-alleles. Dashed line, expected 50:50 eclosion rate in the control condition (no transgene). Number of counted flies (n) are indicated above the bars, χ2-test. c Negative geotaxis assay comparing the motor performance of Drosophila pan-neuronally (nSyb-Gal4) expressing no transgene (Control) or different TyrRS alleles. n ≥ 10 groups of flies. One-way ANOVA with Dunnett Multiple Comparisons test. d NMJ’s of wandering third instar larvae pan-neuronally (nSyb-Gal4) expressing no transgene (Control), TyrRS-WT, TyrRS-WTΔNLS, TyrRS-E196K or TyrRS-E196KΔNLS. HRP, neuronal membrane marker; Dlg1, post-synapse marker. Scale bar, 20 µm. e, f Quantification of total length of the NMJ (e) and total number of boutons (f). n ≥ 10 larvae for each genotype. One-way ANOVA with Dunnett Multiple Comparisons test. g Dye injections reveal the morphology of the giant fiber terminals in flies expressing (R91H05-Gal4) different TyrRS alleles cell autonomously in the giant fibers. Scale bar, 20 µm. h The ability of the giant fiber to follow repeated stimuli at 100 Hz upon presynaptic (R91H05-Gal4) expression of different TyrRS alleles in the giant fiber. n ≥ 20 tergotrochanteral muscles (TTMs) of flies aged to 10 days. One-way ANOVA with Dunnett Multiple Comparisons Test. Box plots show the median, 25–75% percentiles, and 1.5 interquartile range. Source data are provided as a Source Data file