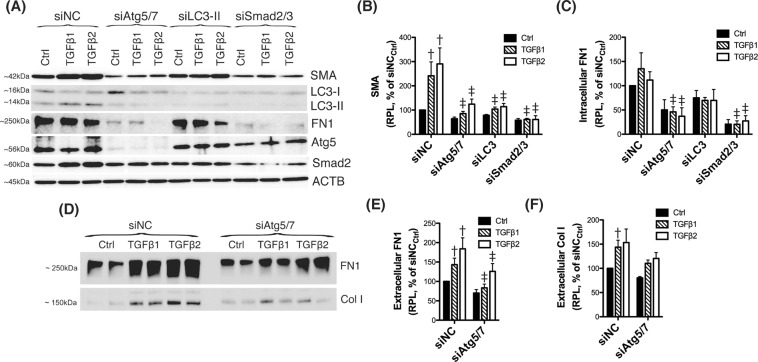
Figure 3.
Silencing autophagy genes inhibits TGFβ-induced fibrosis. Three independent strains of human TM primary cells were transfected with siRNAs targeting Atg5/Atg7 (siAtg5/7), LC3 (siLC3), or Smad2/3 (siSmad2/3). A scrambled siRNA (siNC) was used as control. At 2 d.p.t, cells were treated with either TGFβ1 or TGFβ2 (10 ng/mL) for 48 h. (A) Protein levels of SMA, FN1, Atg5, LC3, and Smad2 evaluated by western blot in whole cell lysates (5 μg). (B) Relative protein levels quantification of the fibrotic markers SMA (B) and FN1 (C) calculated from densitometric analysis of the bands, expressed as percentage of siNCCtrl (set to 100%). β-actin was used as loading control. (D) Protein levels of FN1 and Col I in conditioned-culture media (20 μL) analyzed by western-blot. (E,F) Specific bands were quantified by densitometry and normalized with total protein (μg) of corresponding whole cell lysates, expressed as percentage of siNC. †Denotes statistical significance when comparing TGFβ treatment versus control, using one-way ANOVA with multiple comparisons; ‡denotes statistical significance when comparing TGFβ-treated siAtg5/7, siLC3 or siSmad2 versus their respective siNC treated control, using two-way ANOVA with Bonferroni post-test. Specific mean ± SD and p values, as well as densitometric analysis confirming significant knocked down expression of LC3, Atg5, and Smad2 are included in Supplemental Material. RPL: Relative protein levels.