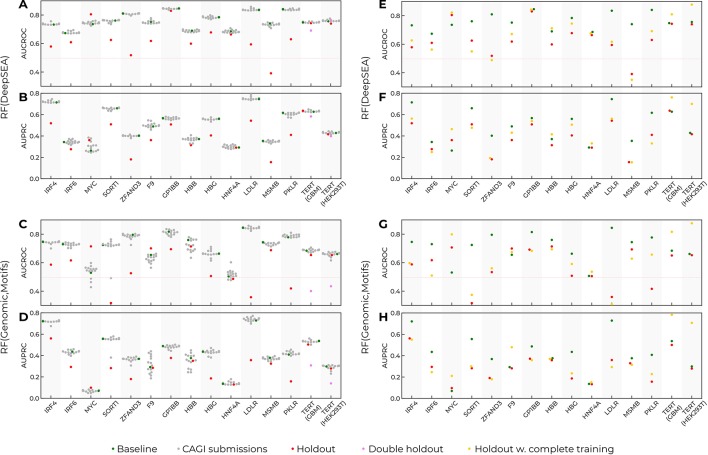
Figure 3.
Prediction performance drops as the data from target reporters are excluded from training. Training on the complete data improves prediction quality, but cannot compensate for the holdout of the data for single-nucleotide variants (SNVs) from the target reporter. Green dots, the baseline models trained in the CAGI setup. Grey dots, the CAGI submissions. Red dots, the models trained in the CAGI setup with the data from the target reporter held out. Violet dots, the performance for the TERT target reporter with the data from both TERT assays held out from training. Yellow dots, the models trained with the complete data from all reporters excluding the target reporter. Reporter names are given at the X-axes. (A, B) Area under precision-recall curve (AUPRC) and area under curve for receiver operating characteristic (AUCROC) for Random Forest with DeepSEA features (baseline and holdout models). (C, D) AUPRC and AUCROC for Random Forest with Genomic signal and sequence motif features (baseline and holdout models). (E, F) AUPRC and AUCROC for Random Forest with DeepSEA features (baseline, holdout, complete training models). (G, H) AUPRC and AUCROC for Random Forest with Genomic signal and sequence motif features (baseline, holdout, complete training models).