Table 2.
Sulfated N-glycan structures derived from glycoproteins of urinary exosomes and detected by either MALDI-MS or LC-MS. Sulfated N-glycans were collected from the aqueous phase of permethylated N-glycans and analyzed with MALDI-MS in the positive ion mode with DHB as a matrix. In addition, reduced native N-glycans fractionated by ion exchange were analyzed with PGC-LC-MS, and some sulfated N-glycans were unequivocally confirmed through MS/MS. H = hexose, N = N-acetylhexosamine, F = deoxyhexose, S = sialic acid, and Su = sulfate group.
| observed m/z MALDI-MS | theoretical m/z MALDI-MS | observed m/z LC-MS | theoretical m/z LC-MS | possible composition | diagnostic fragment ions LC-MS | tentative structure |
|---|---|---|---|---|---|---|
| 1939.7462 (2Na+-H+) | 1939.8793 | 773.3 (949.15*) | 773.3 (948.46*) | H3N4F1Su1 | 486.7, 1059.3, 1342.4 | 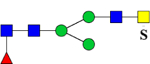 |
| 2143.8398 (2Na+-H+) | 2143.9791 | H4N4F1S1Su1 |
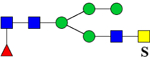 or 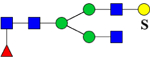
|
|||
| 2173.8552 (2Na+-H+) | 2173.9897 | H5N4Su1 |
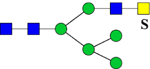 or 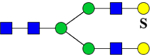
|
|||
| 2348.9128 (2Na+-H+) | 2348.0789 | H5N4F1Su1 | 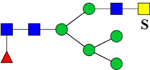 |
|||
| 919.8 (2NH4+) |
902.3 | H5N4Su2 | 445.6, 565.7, 789.9, 1358.3 | 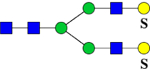 |
||
| 2348.9128 (2Na+-H+) | 2348.0789 | H5N4F1Su1 | 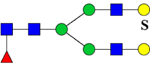 |
|||
| 2388.9534 (2Na+-H+) | 2389.1054 | 1172.41* | 1173.07* | H4N5F1Su1 | 1434.60, 1523.02, 1536.72 | 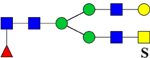 |
| 2436.0403 (3Na+−2H+) | 2436.0020 | 984.5 (NH4+) |
975.3 | H5N4F1Su2 | 445.6, 607.8, 854.1, 1342.2 | 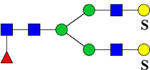 |
| 2477.0867 (3Na+−2H+) | 2477.0285 | 1206.82* | 1206.05* | H4N5F1Su2 | 942.24, 1058.98, 1756.61 | 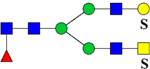 |
| 2517.8914 (3Na+−2H+) | 2518.0551 | H3N6F1Su2 | 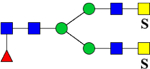 |
|||
| 2629.2751 (2Na+-H+) | 2629.1358 | 1294.37* | 1293.09* | H4N5F2Su2 | 547.69, 778.89, 1059.85, 1655.25 | 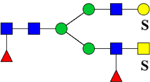 |
| 1008.6 | 1007.8 | H5N4S1Su1 | 365.6, 445.6, 656.9, 1358.3, 1569.3 | |||
| 1029.2** | 1028.4 | H4N4S1Su1 | 486.6, 656.9, 1569.4, 1852.1 | |||
| 1029.0** | 486.7, 657.0, 882.6, 1569.3, | |||||
| 2711.1006 (3Na+−2H+) | 2711.1389 | 1094.5 (NH4+) |
1084.8 | H6N5Su2 | 445.6, 973.2, 1196.2, 1803.1 | 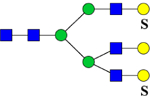 |
| 2728.2505 (Na+) | 2728.2971 | 1110.9** (NH4+) |
1101.4 | H4N4F1S1Su1 | 365.6, 656.9, 956.1 | 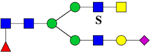 |
| 1110.8** (NH4+) |
365.6, 657.0, 955.9, 1545.3 | |||||
| 2670.2996 (3Na+−2H+) | 2670.1123 | H7N4Su2 | 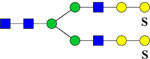 |
|||
| 2670.2996 (3Na+−2H+) | 2670.1623 | H3N6F2Su2 | 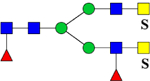 |
|||
| 2758.2343 (Na+) | 2758.3077 | H5N5S1Su1 | 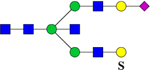 |
|||
| 2824.3269 (Na+) | 2824.2489 | H5N5S1Su2 | 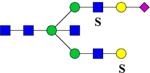 |
|||
| 1191.5 | 1190.4 | H6N5S1Su1 | 365.5, 445.6, 657.0, 968.3, 2014.2 | 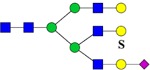 |
||
| 1130.7 | 1129.9 | H4N6S1Su1 | 656.9, 860.1, 1399.2, 2055.2 | 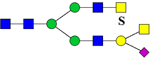 |
||
| 3166.3882 (Na+) |
3166.5072 | H7N5SlSul | 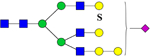 |
|||
| 3166.3882 (Na+) |
3166.3779 | H6N5S2Su2 | ||||
| 945.8 (NH4+, +3) |
939.7 (+3) | H5N6F1S2Su1 | 445.4, 657.0, 1080.7, 1186.7 | 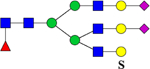 |
||
| 3066.5334 (2Na+-H+) | 3066.4425 | H4N7S1Su1 | 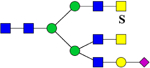 |
|||
| 2880.3503 (Na+) | 2880.4714 | H4N6F3(Su) | 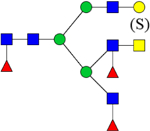 |
Sulfated N-glycans released through chloroform extraction after the permethylation step
Multiple peaks of the same possible composition were detected during chromatography
